MicroRNA-33a mediates the regulation of high mobility group AT-hook 2 gene (HMGA2) by thyroid transcription factor 1 (TTF-1/NKX2-1)
- PMID: 23625920
- PMCID: PMC3675572
- DOI: 10.1074/jbc.M113.474643
MicroRNA-33a mediates the regulation of high mobility group AT-hook 2 gene (HMGA2) by thyroid transcription factor 1 (TTF-1/NKX2-1)
Abstract
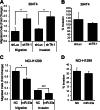
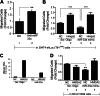
In lung cancers, TTF-1 displays seemingly paradoxical activities. Although TTF-1 is amplified in primary human lung cancers, it inhibits primary lung tumors from metastasizing in a mouse model system. It was reported that the oncogenic proepithelial mesenchymal transition (EMT) high mobility group AT-hook 2 gene (HMGA2) mediates the antimetastatic function of TTF-1. To gain mechanistic insight into the metastasis-critical signaling axis of TTF-1 to HMGA2, we used both reverse and forward strategies and discovered that microRNA-33a (miR-33a) is under direct positive regulation of TTF-1. By chromatin immunoprecipitation, we determined that TTF-1 binds to the promoter of SREBF2, the host gene of miR-33a. The 3'-untranslated region (UTR) of HMGA2 contains three predicted binding sites of miR-33a. We showed that the first two highly conserved sites are conducive to HMGA2 repression by miR-33a, establishing HMGA2 as a genuine target of miR-33a. Functional studies revealed that enforced expression of miR-33a inhibits the motility of lung cancer cells, and this inhibition can be rescued by overexpression of the form of HMGA2 without the 3'-UTR, suggesting that TTF-1 keeps the prometastasis gene HMGA2 in check via up-regulating miR-33a. This study reports the first miRNAs directly regulated by TTF-1 and clarifies how TTF-1 controls HMGA2 expression. Moreover, the documented importance of SREBF2 and miR-33a in regulating cholesterol metabolism suggests that TTF-1 may be a modulator of cholesterol homeostasis in the lung. Future studies will be dedicated to understanding how miRNAs influence the oncogenic activity of TTF-1 and the role of TTF-1 in cholesterol metabolism.
Keywords: Cholesterol Regulation; Gene Regulation; HMGA2; Lung Cancer; Metastasis; MicroRNA; SREBP2; TTF-1; miR-32; miR-33a.
Figures



Similar articles
-
MiR-365 regulates lung cancer and developmental gene thyroid transcription factor 1.Cell Cycle. 2012 Jan 1;11(1):177-86. doi: 10.4161/cc.11.1.18576. Epub 2012 Jan 1. Cell Cycle. 2012. PMID: 22185756 Free PMC article.
-
Transcription factor and microRNA interactions in lung cells: an inhibitory link between NK2 homeobox 1, miR-200c and the developmental and oncogenic factors Nfib and Myb.Respir Res. 2015 Feb 13;16(1):22. doi: 10.1186/s12931-015-0186-6. Respir Res. 2015. PMID: 25763778 Free PMC article.
-
Downregulation of long noncoding RNA CRNDE suppresses drug resistance of liver cancer cells by increasing microRNA-33a expression and decreasing HMGA2 expression.Cell Cycle. 2019 Oct;18(19):2524-2537. doi: 10.1080/15384101.2019.1652035. Epub 2019 Aug 15. Cell Cycle. 2019. Retraction in: Cell Cycle. 2023 Jul-Aug;22(14-16):1801. doi: 10.1080/15384101.2023.2211862. PMID: 31416393 Free PMC article. Retracted.
-
MicroRNAs regulating lipid metabolism in atherogenesis.Thromb Haemost. 2012 Apr;107(4):642-7. doi: 10.1160/TH11-10-0694. Epub 2012 Jan 25. Thromb Haemost. 2012. PMID: 22274626 Free PMC article. Review.
-
Thyroid transcription factor-1 (TTF-1/Nkx2.1/TITF1) gene regulation in the lung.Clin Sci (Lond). 2009 Jan;116(1):27-35. doi: 10.1042/CS20080068. Clin Sci (Lond). 2009. PMID: 19037882 Review.
Cited by
-
Paired Box 9 (PAX9), the RNA polymerase II transcription factor, regulates human ribosome biogenesis and craniofacial development.PLoS Genet. 2020 Aug 19;16(8):e1008967. doi: 10.1371/journal.pgen.1008967. eCollection 2020 Aug. PLoS Genet. 2020. PMID: 32813698 Free PMC article.
-
Connecting Cholesterol Efflux Factors to Lung Cancer Biology and Therapeutics.Int J Mol Sci. 2021 Jul 5;22(13):7209. doi: 10.3390/ijms22137209. Int J Mol Sci. 2021. PMID: 34281263 Free PMC article. Review.
-
MircoRNA-33a inhibits epithelial-to-mesenchymal transition and metastasis and could be a prognostic marker in non-small cell lung cancer.Sci Rep. 2015 Sep 2;5:13677. doi: 10.1038/srep13677. Sci Rep. 2015. Retraction in: Sci Rep. 2021 Apr 14;11(1):8536. doi: 10.1038/s41598-021-88178-8. PMID: 26330060 Free PMC article. Retracted.
-
Thyroid transcription factor-1-regulated microRNA-532-5p targets KRAS and MKL2 oncogenes and induces apoptosis in lung adenocarcinoma cells.Cancer Sci. 2017 Jul;108(7):1394-1404. doi: 10.1111/cas.13271. Epub 2017 Jun 10. Cancer Sci. 2017. PMID: 28474808 Free PMC article.
-
miR-33a is downregulated in melanoma cells and modulates cell proliferation by targeting PCTAIRE1.Oncol Lett. 2016 Apr;11(4):2741-2746. doi: 10.3892/ol.2016.4321. Epub 2016 Mar 9. Oncol Lett. 2016. PMID: 27073545 Free PMC article.
References
-
- Bartel D. P. (2004) MicroRNAs. Genomics, Biogenesis, Mechanism, and Function. Cell 116, 281–297 - PubMed
-
- Esquela-Kerscher A., Slack F. J. (2006) Oncomirs. MicroRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269 - PubMed
-
- He L., Hannon G. J. (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

