Small-molecule inhibitors of acetyltransferase p300 identified by high-throughput screening are potent anticancer agents
- PMID: 23625935
- PMCID: PMC3651759
- DOI: 10.1158/1535-7163.MCT-12-0930
Small-molecule inhibitors of acetyltransferase p300 identified by high-throughput screening are potent anticancer agents
Erratum in
- Mol Cancer Ther. 2013 Aug;12(8):1688. Chase, Peter [added]
Abstract
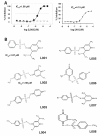
Acetyltransferase p300 (KAT3B) plays key roles in signaling cascades that support cancer cell survival and sustained proliferation. Thus, p300 represents a potential anticancer therapeutic target. To discover novel anticancer agents that target p300, we conducted a high-throughput screening campaign. A library of 622,079 compounds was assayed for cytotoxicity to the triple-negative breast cancer (TNBC) cell line MDA-MB-231 but not to the human mammary epithelial cells. The resulting compounds were tested in a biochemical assay for inhibiting the enzymatic activity of p300. One compound (L002, NSC764414) displayed an IC50 of 1.98 μmol/L against p300 in vitro, inhibited acetylation of histones and p53, and suppressed STAT3 activation in cell-based assays. L002 could be docked to the active site of the p300 catalytic domain. Biochemical tests of a series of related compounds revealed functional groups that may impact inhibitory potency of L002 against p300. Interestingly, these analogs showed inhibitory activities against the cellular paralog of p300 (CBP), p300/CBP-associated factor, and GCN5, but not to other acetyltransferases (KAT5, KAT6B, and KAT7), histone deacetylases, and histone methyltransferases. Among the NCI-60 panel of cancer cell lines, leukemia and lymphoma cell lines were extremely sensitive to L002, whereas it is toxic to only a limited number of cell lines derived from solid tumors. Notably, breast cancer cell lines, especially those derived from TNBC, were highly susceptible to L002. In vivo, it potently suppressed tumor growth and histone acetylation of MDA-MB-468 xenografts. Thus, these new acetyltransferase inhibitors are potential anticancer therapeutics.
©2013 AACR
Figures

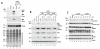



References
-
- Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, et al. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–84. - PubMed
-
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

