Explaining microbial phenotypes on a genomic scale: GWAS for microbes
- PMID: 23625995
- PMCID: PMC3743258
- DOI: 10.1093/bfgp/elt008
Explaining microbial phenotypes on a genomic scale: GWAS for microbes
Abstract
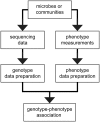
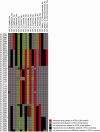
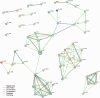
There is an increasing availability of complete or draft genome sequences for microbial organisms. These data form a potentially valuable resource for genotype-phenotype association and gene function prediction, provided that phenotypes are consistently annotated for all the sequenced strains. In this review, we address the requirements for successful gene-trait matching. We outline a basic protocol for microbial functional genomics, including genome assembly, annotation of genotypes (including single nucleotide polymorphisms, orthologous groups and prophages), data pre-processing, genotype-phenotype association, visualization and interpretation of results. The methodologies for association described herein can be applied to other data types, opening up possibilities to analyze transcriptome-phenotype associations, and correlate microbial population structure or activity, as measured by metagenomics, to environmental parameters.
Keywords: functional genomics; genome-wide association studies; genotype–phenotype association; microbial genomics; random forest.
Figures



References
-
- Bork P, Dandekar T, Diaz-Lazcoz Y, et al. Predicting function: from genes to genomes and back. J Mol Biol. 1998;283:707–25. - PubMed
-
- Korbel JO, Jensen LJ, von Mering C, et al. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat Biotechnol. 2004;22:911–7. - PubMed
-
- Kensche PR, Oti M, Dutilh BE, et al. Conservation of divergent transcription in fungi. Trends Genet. 2008;24:207–11. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

