Metagenomic evidence for sulfur lithotrophy by Epsilonproteobacteria as the major energy source for primary productivity in a sub-aerial arctic glacial deposit, Borup Fiord Pass
- PMID: 23626586
- PMCID: PMC3631710
- DOI: 10.3389/fmicb.2013.00063
Metagenomic evidence for sulfur lithotrophy by Epsilonproteobacteria as the major energy source for primary productivity in a sub-aerial arctic glacial deposit, Borup Fiord Pass
Abstract
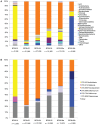
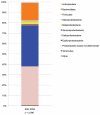
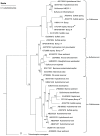
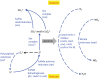
We combined free enenergy calculations and metagenomic analyses of an elemental sulfur (S(0)) deposit on the surface of Borup Fiord Pass Glacier in the Canadian High Arctic to investigate whether the energy available from different redox reactions in an environment predicts microbial metabolism. Many S, C, Fe, As, Mn, and [Formula: see text] oxidation reactions were predicted to be energetically feasible in the deposit, and aerobic oxidation of S(0) was the most abundant chemical energy source. Small subunit ribosomal RNA (SSU rRNA) gene sequence data showed that the dominant phylotypes were Sulfurovum and Sulfuricurvum, both Epsilonproteobacteria known to be capable of sulfur lithotrophy. Sulfur redox genes were abundant in the metagenome, but sox genes were significantly more abundant than reverse dsr (dissimilatory sulfite reductase)genes. Interestingly, there appeared to be habitable niches that were unoccupied at the depth of genome coverage obtained. Photosynthesis and [Formula: see text] oxidation should both be energetically favorable, but we found few or no functional genes for oxygenic or anoxygenic photosynthesis, or for [Formula: see text] oxidation by either oxygen (nitrification) or nitrite (anammox). The free energy, SSU rRNA gene and quantitative functional gene data are all consistent with the hypothesis that sulfur-based chemolithoautotrophy by Epsilonproteobacteria (Sulfurovum and Sulfuricurvum) is the main form of primary productivity at this site, instead of photosynthesis. This is despite the presence of 24-h sunlight, and the fact that photosynthesis is not known to be inhibited by any of the environmental conditions present. This is the first time that Sulfurovum and Sulfuricurvum have been shown to dominate a sub-aerial environment, rather than anoxic or sulfidic settings. We also found that Flavobacteria dominate the surface of the sulfur deposits. We hypothesize that this aerobic heterotroph uses enough oxygen to create a microoxic environment in the sulfur below, where the Epsilonproteobacteria can flourish.
Keywords: Epsilonproteobacteria; Sulfurovum; arctic; free energy; lithotrophy; metagenome; photosynthesis; sulfur.
Figures




Similar articles
-
Low-Temperature Sulfidic-Ice Microbial Communities, Borup Fiord Pass, Canadian High Arctic.Front Microbiol. 2018 Jul 24;9:1622. doi: 10.3389/fmicb.2018.01622. eCollection 2018. Front Microbiol. 2018. PMID: 30087659 Free PMC article.
-
Microbial Metabolic Redundancy Is a Key Mechanism in a Sulfur-Rich Glacial Ecosystem.mSystems. 2020 Aug 4;5(4):e00504-20. doi: 10.1128/mSystems.00504-20. mSystems. 2020. PMID: 32753510 Free PMC article.
-
Metagenomic insights into S(0) precipitation in a terrestrial subsurface lithoautotrophic ecosystem.Front Microbiol. 2015 Jan 8;5:756. doi: 10.3389/fmicb.2014.00756. eCollection 2014. Front Microbiol. 2015. PMID: 25620962 Free PMC article.
-
Phylogenetic diversity and functional gene patterns of sulfur-oxidizing subseafloor Epsilonproteobacteria in diffuse hydrothermal vent fluids.Front Microbiol. 2013 Jul 8;4:185. doi: 10.3389/fmicb.2013.00185. eCollection 2013. Front Microbiol. 2013. PMID: 23847608 Free PMC article.
-
Hydrogen sulfide, reactive sulfur species and coping with reactive oxygen species.Free Radic Biol Med. 2019 Aug 20;140:74-83. doi: 10.1016/j.freeradbiomed.2019.01.020. Epub 2019 Jan 28. Free Radic Biol Med. 2019. PMID: 30703482 Review.
Cited by
-
Microbial carbon metabolism associated with electrogenic sulphur oxidation in coastal sediments.ISME J. 2015 Sep;9(9):1966-78. doi: 10.1038/ismej.2015.10. Epub 2015 Feb 13. ISME J. 2015. PMID: 25679534 Free PMC article.
-
Low-Temperature Sulfidic-Ice Microbial Communities, Borup Fiord Pass, Canadian High Arctic.Front Microbiol. 2018 Jul 24;9:1622. doi: 10.3389/fmicb.2018.01622. eCollection 2018. Front Microbiol. 2018. PMID: 30087659 Free PMC article.
-
Sulfur-cycling chemolithoautotrophic microbial community dominates a cold, anoxic, hypersaline Arctic spring.Microbiome. 2023 Sep 11;11(1):203. doi: 10.1186/s40168-023-01628-5. Microbiome. 2023. PMID: 37697305 Free PMC article.
-
Disproportionation of Inorganic Sulfur Compounds by Mesophilic Chemolithoautotrophic Campylobacterota.mSystems. 2023 Feb 23;8(1):e0095422. doi: 10.1128/msystems.00954-22. Epub 2022 Dec 21. mSystems. 2023. PMID: 36541763 Free PMC article.
-
Geomicrobiology of a seawater-influenced active sulfuric acid cave.PLoS One. 2019 Aug 8;14(8):e0220706. doi: 10.1371/journal.pone.0220706. eCollection 2019. PLoS One. 2019. PMID: 31393920 Free PMC article.
References
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410 - PubMed
-
- Amend J. P., McCollom T. M., Hentscher M., Bach W. (2011). Catabolic and anabolic energy for chemolithoautotrophs in deep-sea hydrothermal systems hosted in different rock types. Geochim. Cosmochim. Acta 75 5736–5748
-
- Amend J. P., Shock E. L. (2001). Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol. Rev. 25 175–243 - PubMed
-
- Borin S., Brusetti L., Daffonchio D., Delaney E., Baldi F. (2009). Biodiversity of prokaryotic communities in sediments of different sub-basins of the Venice lagoon. Res. Microbiol. 160 307–314 - PubMed
-
- Boyd E., Hamilton T. L., Spear J. R., Lavin M., Peters J. W. (2010). [FeFe]-hydrogenase in Yellowstone National Park: evidence for dispersal limitation and phylogenetic niche conservatism. ISME J. 4 1485–1495 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

