Filter characteristics influencing circulating tumor cell enrichment from whole blood
- PMID: 23626725
- PMCID: PMC3634026
- DOI: 10.1371/journal.pone.0061770
Filter characteristics influencing circulating tumor cell enrichment from whole blood
Abstract
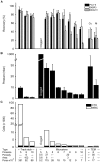
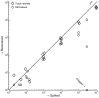
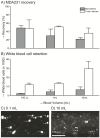
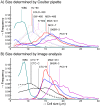
A variety of filters assays have been described to enrich circulating tumor cells (CTC) based on differences in physical characteristics of blood cells and CTC. In this study we evaluate different filter types to derive the properties of the ideal filter for CTC enrichment. Between 0.1 and 10 mL of whole blood spiked with cells from tumor cell lines were passed through silicon nitride microsieves, polymer track-etched filters and metal TEM grids with various pore sizes. The recovery and size of 9 different culture cell lines was determined and compared to the size of EpCAM+CK+CD45-DNA+ CTC from patients with metastatic breast, colorectal and prostate cancer. The 8 µm track-etched filter and the 5 µm microsieve had the best performance on MDA-231, PC3-9 and SKBR-3 cells, enriching >80% of cells from whole blood. TEM grids had poor recovery of ∼25%. Median diameter of cell lines ranged from 10.9-19.0 µm, compared to 13.1, 10.7, and 11.0 µm for breast, prostate and colorectal CTC, respectively. The 11.4 µm COLO-320 cell line had the lowest recovery of 17%. The ideal filter for CTC enrichment is constructed of a stiff, flat material, is inert to blood cells, has at least 100,000 regularly spaced 5 µm pores for 1 ml of blood with a ≤10% porosity. While cell size is an important factor in determining recovery, other factors must be involved as well. To evaluate a filtration procedure, cell lines with a median size of 11-13 µm should be used to challenge the system.
Conflict of interest statement
Figures


References
-
- Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, et al. (2004) Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 351: 781–791. - PubMed
-
- Pierga JY, Hajage D, Bachelot T, Delaloge S, Brain E, et al. (2012) High independent prognostic and predictive value of circulating tumor cells compared with serum tumor markers in a large prospective trial in first-line chemotherapy for metastatic breast cancer patients. Ann Oncol 23: 618–24. - PubMed
-
- Tol J, Koopman M, Miller MC, Tibbe A, Cats A, et al. (2010) Circulating tumour cells early predict progression-free and overall survival in advanced colorectal cancer patients treated with chemotherapy and targeted agents. Ann Oncol 21: 1006–1012. - PubMed
-
- de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, et al. (2008) Circulating Tumor Cells Predict Survival Benefit from Treatment in Metastatic Castration-Resistant Prostate Cancer. Clin Cancer Res 14: 6302–6309. - PubMed
-
- Rao C, Bui T, Connelly M, Doyle G, Karydis I, et al. (2011) Circulating melanoma cells and survival in metastatic melanoma. Int J Oncol 38: 755–760. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

