Development of GMDR-GPU for gene-gene interaction analysis and its application to WTCCC GWAS data for type 2 diabetes
- PMID: 23626757
- PMCID: PMC3633958
- DOI: 10.1371/journal.pone.0061943
Development of GMDR-GPU for gene-gene interaction analysis and its application to WTCCC GWAS data for type 2 diabetes
Abstract
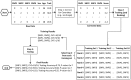
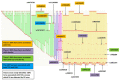
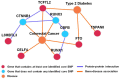
Although genome-wide association studies (GWAS) have identified a significant number of single-nucleotide polymorphisms (SNPs) associated with many complex human traits, the susceptibility loci identified so far can explain only a small fraction of the genetic risk. Among other possible explanations, the lack of a comprehensive examination of gene-gene interaction (G×G) is often considered a source of the missing heritability. Previously, we reported a model-free Generalized Multifactor Dimensionality Reduction (GMDR) approach for detecting G×G in both dichotomous and quantitative phenotypes. However, the computational burden and less efficient implementation of the original programs make them impossible to use for GWAS. In this study, we developed a graphics processing unit (GPU)-based GMDR program (named GWAS-GPU), which is able not only to analyze GWAS data but also to run much faster than the earlier version of the GMDR program. As a demonstration of the program, we used the GMDR-GPU software to analyze a publicly available GWAS dataset on type 2 diabetes (T2D) from the Wellcome Trust Case Control Consortium. Through an exhaustive search of pair-wise interactions and a selected search of three- to five-way interactions conditioned on significant pair-wise results, we identified 24 core SNPs in six genes (FTO: rs9939973, rs9940128, rs9922047, rs1121980, rs9939609, rs9930506; TSPAN8: rs1495377; TCF7L2: rs4074720, rs7901695, rs4506565, rs4132670, rs10787472, rs11196205, rs10885409, rs11196208; L3MBTL3: rs10485400, rs4897366; CELF4: rs2852373, rs608489; RUNX1: rs445984, rs1040328, rs990074, rs2223046, rs2834970) that appear to be important for T2D. Of these core SNPs, 11 in FTO, TSPAN8, and TCF7L2 have been reported to be associated with T2D, obesity, or both, providing an independent replication of previously reported SNPs. Importantly, we identified three new susceptibility genes; i.e., L3MBTL3, CELF4, and RUNX1, for T2D, a finding that warrants further investigation with independent samples.
Conflict of interest statement
Figures
Similar articles
-
Replication and Relevance of Multiple Susceptibility Loci Discovered from Genome Wide Association Studies for Type 2 Diabetes in an Indian Population.PLoS One. 2016 Jun 16;11(6):e0157364. doi: 10.1371/journal.pone.0157364. eCollection 2016. PLoS One. 2016. PMID: 27310578 Free PMC article.
-
GWIS--model-free, fast and exhaustive search for epistatic interactions in case-control GWAS.BMC Genomics. 2013;14 Suppl 3(Suppl 3):S10. doi: 10.1186/1471-2164-14-S3-S10. Epub 2013 May 28. BMC Genomics. 2013. PMID: 23819779 Free PMC article.
-
Gene, pathway and network frameworks to identify epistatic interactions of single nucleotide polymorphisms derived from GWAS data.BMC Syst Biol. 2012;6 Suppl 3(Suppl 3):S15. doi: 10.1186/1752-0509-6-S3-S15. Epub 2012 Dec 17. BMC Syst Biol. 2012. PMID: 23281810 Free PMC article.
-
Shared genetic etiology underlying Alzheimer's disease and type 2 diabetes.Mol Aspects Med. 2015 Jun-Oct;43-44:66-76. doi: 10.1016/j.mam.2015.06.006. Epub 2015 Jun 23. Mol Aspects Med. 2015. PMID: 26116273 Free PMC article. Review.
-
Epistasis, complexity, and multifactor dimensionality reduction.Methods Mol Biol. 2013;1019:465-77. doi: 10.1007/978-1-62703-447-0_22. Methods Mol Biol. 2013. PMID: 23756906 Review.
Cited by
-
A roadmap to multifactor dimensionality reduction methods.Brief Bioinform. 2016 Mar;17(2):293-308. doi: 10.1093/bib/bbv038. Epub 2015 Jun 24. Brief Bioinform. 2016. PMID: 26108231 Free PMC article. Review.
-
Evolutionary footprint of epistasis.PLoS Comput Biol. 2018 Sep 17;14(9):e1006426. doi: 10.1371/journal.pcbi.1006426. eCollection 2018 Sep. PLoS Comput Biol. 2018. PMID: 30222748 Free PMC article.
-
Dissection of genetic architecture of rice plant height and heading date by multiple-strategy-based association studies.Sci Rep. 2016 Jul 13;6:29718. doi: 10.1038/srep29718. Sci Rep. 2016. PMID: 27406081 Free PMC article.
-
Genome-wide association study of maize plant architecture using F1 populations.Plant Mol Biol. 2019 Jan;99(1-2):1-15. doi: 10.1007/s11103-018-0797-7. Epub 2018 Dec 5. Plant Mol Biol. 2019. PMID: 30519826
-
Dissection of complicate genetic architecture and breeding perspective of cottonseed traits by genome-wide association study.BMC Genomics. 2018 Jun 13;19(1):451. doi: 10.1186/s12864-018-4837-0. BMC Genomics. 2018. PMID: 29895260 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

