Interactions among quorum sensing inhibitors
- PMID: 23626795
- PMCID: PMC3633908
- DOI: 10.1371/journal.pone.0062254
Interactions among quorum sensing inhibitors
Abstract
Many pathogenic bacteria use quorum sensing (QS) systems to regulate the expression of virulence genes in a density-dependent manner. In one widespread QS paradigm the enzyme LuxI generates a small diffusible molecule of the acyl-homoserine lactone (AHL) family; high cell densities lead to high AHL levels; AHL binds the transcription factor LuxR, triggering it to activate gene expression at a virulence promoter. The emergence of antibiotic resistance has generated interest in alternative anti-microbial therapies that target QS. Inhibitors of LuxI and LuxR have been developed and tested in vivo, and can act at various levels: inhibiting the synthesis of AHL by LuxI, competitively or non-competitively inhibiting LuxR, or increasing the turnover of LuxI, LuxR, or AHL. Here use an experimentally validated computational model of LuxI/LuxR QS to study the effects of using inhibitors individually and in combination. The model includes the effect of transcriptional feedback, which generates highly non-linear responses as inhibitor levels are increased. For the ubiquitous LuxI-feedback virulence systems, inhibitors of LuxI are more effective than those of LuxR when used individually. Paradoxically, we find that LuxR competitive inhibitors, either individually or in combination with other inhibitors, can sometimes increase virulence by weakly activating LuxR. For both LuxI-feedback and LuxR-feedback systems, a combination of LuxR non-competitive inhibitors and LuxI inhibitors act multiplicatively over a broad parameter range. In our analysis, this final strategy emerges as the only robust therapeutic option.
Conflict of interest statement
Figures

 ; dot-dashed); with a LuxR non-competitive inhibitor (
; dot-dashed); with a LuxR non-competitive inhibitor ( ; dashed); and with a LuxR competitive inhibitor (
; dashed); and with a LuxR competitive inhibitor ( ; dotted). Positive feedback produces induction curves with stable upper and lower branches separated by an unstable middle branch. Cells which start off on the un-induced lower branch will jump to the highly-induced upper branch when their density crosses a critical threshold. (C) LuxI-feedback system. (D) LuxR-feedback system.
; dotted). Positive feedback produces induction curves with stable upper and lower branches separated by an unstable middle branch. Cells which start off on the un-induced lower branch will jump to the highly-induced upper branch when their density crosses a critical threshold. (C) LuxI-feedback system. (D) LuxR-feedback system.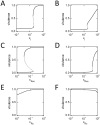
 in the absence of inhibitors,
in the absence of inhibitors,  at high inhibitor levels). Parameter values are taken from Table 1. The x-axis is logarithmic. Due to positive feedback, the equations sometimes admit three solutions for a fixed level of inhibition: the upper and lower branches are stable (solid curves); the middle branch is unstable (dotted curve). (A,C,E) LuxI-feedback systems with α = 0.11. (B,D,F) LuxR-feedback systems with α = 0.05. (A,B) LuxI inhibitors (varying
at high inhibitor levels). Parameter values are taken from Table 1. The x-axis is logarithmic. Due to positive feedback, the equations sometimes admit three solutions for a fixed level of inhibition: the upper and lower branches are stable (solid curves); the middle branch is unstable (dotted curve). (A,C,E) LuxI-feedback systems with α = 0.11. (B,D,F) LuxR-feedback systems with α = 0.05. (A,B) LuxI inhibitors (varying  ). (C,D) LuxR non-competitive inhibitors (varying
). (C,D) LuxR non-competitive inhibitors (varying  ). (E,F) LuxR competitive inhibitors (varying
). (E,F) LuxR competitive inhibitors (varying  ).
).

 close to 1), the interaction with the non-competitive inhibitor is antagonistic. For higher values of competitive inhibition (
close to 1), the interaction with the non-competitive inhibitor is antagonistic. For higher values of competitive inhibition ( close to 0) the interaction with the non-competitive inhibitor is multiplicative. The dark curve in {α,n} space separates purely abrupt responses from a mixture of smooth and abrupt responses. (D) Above the value α = 0.125, both the LuxR competitive and non-competitive inhibitors act to suppress virulence. However, it is mainly the level of the LuxR non-competitive inhibitor which is important. Below the value α = 0.125, the competitive inhibitor acts antagonistically with the non-competitive inhibitor. For n >1.4 the response is abrupt; for n <1.4 the response is smooth. (E) The two inhibitors show similar interactions as in panel (C). The dark curves separate regions where the response is completely smooth (top), completely abrupt (bottom left) or a mixture of the two (bottom right). (F) Virulence expression is high over all inhibitor combinations and parameter values shown. The distinction between cooperative and antagonistic behavior is hardly visible.
close to 0) the interaction with the non-competitive inhibitor is multiplicative. The dark curve in {α,n} space separates purely abrupt responses from a mixture of smooth and abrupt responses. (D) Above the value α = 0.125, both the LuxR competitive and non-competitive inhibitors act to suppress virulence. However, it is mainly the level of the LuxR non-competitive inhibitor which is important. Below the value α = 0.125, the competitive inhibitor acts antagonistically with the non-competitive inhibitor. For n >1.4 the response is abrupt; for n <1.4 the response is smooth. (E) The two inhibitors show similar interactions as in panel (C). The dark curves separate regions where the response is completely smooth (top), completely abrupt (bottom left) or a mixture of the two (bottom right). (F) Virulence expression is high over all inhibitor combinations and parameter values shown. The distinction between cooperative and antagonistic behavior is hardly visible.References
-
- Neu HC (1992) The crisis in antibiotic resistance. Science 257: 1064–1073. - PubMed
-
- Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405: 299–304. - PubMed
-
- Livermore DM (2003) Bacterial resistance: origins, epidemiology, and impact. Clin Infect Dis 36: S11–S23. - PubMed
-
- Rybtke MT, Jensen PØ, Høiby N, Givskov M, Tolker-Nielsen T, et al. (2011) The implication of Pseudomonas aeruginosa biofilms in infections. Inflamm Allergy Drug Targets 10: 141–157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

