Single-stranded DNA as a cleavable linker for bioorthogonal click chemistry-based proteomics
- PMID: 23627610
- PMCID: PMC4078914
- DOI: 10.1021/bc400093x
Single-stranded DNA as a cleavable linker for bioorthogonal click chemistry-based proteomics
Abstract
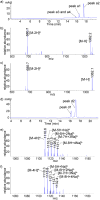
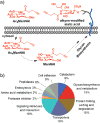
In this communication, we report a new class of cleavable linker based on automatically synthesized, single-stranded DNAs. We incorporated a DNA oligo into an azide-functionalized biotin (biotin-DNA-N3) and used the probe to enrich for alkyne-tagged glycoproteins from mammalian cell lysates. Highly efficient and selective release of the captured proteins from streptavidin agarose resins was achieved using DNase treatment under very mild conditions. A total of 36 sialylated glycoproteins were identified from the lysates of HL60 cells, an acute human promyeloid leukemia cell line. These sialylated glycoproteins were involved in many different biological processes ranging from glycan biosynthesis to cell adhesion events.
Conflict of interest statement
Figures



References
-
- Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed. 2002;41:2596–2599. - PubMed
-
- Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem. 2002;67:3057–3064. - PubMed
-
- Speers AE, Cravatt BF. Chemical strategies for activity-based proteomics. ChemBioChem. 2004;5:41–47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

