Charcot-Marie-Tooth disease-associated mutants of GDAP1 dissociate its roles in peroxisomal and mitochondrial fission
- PMID: 23628762
- PMCID: PMC3674444
- DOI: 10.1038/embor.2013.56
Charcot-Marie-Tooth disease-associated mutants of GDAP1 dissociate its roles in peroxisomal and mitochondrial fission
Abstract
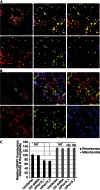
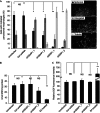
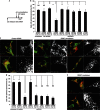
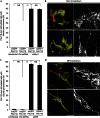
Mitochondria and peroxisomes can be fragmented by the process of fission. The fission machineries of both organelles share a set of proteins. GDAP1 is a tail-anchored protein of mitochondria and induces mitochondrial fragmentation. Mutations in GDAP1 lead to Charcot-Marie-Tooth disease (CMT), an inherited peripheral neuropathy, and affect mitochondrial dynamics. Here, we show that GDAP1 is also targeted to peroxisomes mediated by the import receptor Pex19. Knockdown of GDAP1 leads to peroxisomal elongation that can be rescued by re-expressing GDAP1 and by missense mutated forms found in CMT patients. GDAP1-induced peroxisomal fission is dependent on the integrity of its hydrophobic domain 1, and on Drp1 and Mff, as is mitochondrial fission. Thus, GDAP1 regulates mitochondrial and peroxisomal fission by a similar mechanism. However, our results reveal also a more critical role of the amino-terminal GDAP1 domains, carrying most CMT-causing mutations, in the regulation of mitochondrial compared to peroxisomal fission.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
-
- Wanders RJ, Waterham HR (2006) Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem 75: 295–332 - PubMed
-
- Fagarasanu A, Fagarasanu M, Rachubinski RA (2007) Maintaining peroxisome populations: a story of division and inheritance. Annu Rev Cell Dev Biol 23: 321–344 - PubMed
-
- Schrader M, Fahimi HD (2006) Growth and division of peroxisomes. Int Rev Cytol 255: 237–290 - PubMed
-
- van der Zand A, Gent J, Braakman I, Tabak HF (2012) Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 149: 397–409 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

