Improved nutritive quality and salt resistance in transgenic maize by simultaneously overexpression of a natural lysine-rich protein gene, SBgLR, and an ERF transcription factor gene, TSRF1
- PMID: 23629675
- PMCID: PMC3676793
- DOI: 10.3390/ijms14059459
Improved nutritive quality and salt resistance in transgenic maize by simultaneously overexpression of a natural lysine-rich protein gene, SBgLR, and an ERF transcription factor gene, TSRF1
Abstract
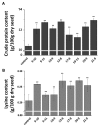
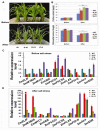
Maize (Zea mays L.), as one of the most important crops in the world, is deficient in lysine and tryptophan. Environmental conditions greatly impact plant growth, development and productivity. In this study, we used particle bombardment mediated co-transformation to obtain marker-free transgenic maize inbred X178 lines harboring a lysine-rich protein gene SBgLR from potato and an ethylene responsive factor (ERF) transcription factor gene, TSRF1, from tomato. Both of the target genes were successfully expressed and showed various expression levels in different transgenic lines. Analysis showed that the protein and lysine content in T1 transgenic maize seeds increased significantly. Compared to non-transformed maize, the protein and lysine content increased by 7.7% to 24.38% and 8.70% to 30.43%, respectively. Moreover, transgenic maize exhibited more tolerance to salt stress. When treated with 200 mM NaCl for 48 h, both non-transformed and transgenic plant leaves displayed wilting and losing green symptoms and dramatic increase of the free proline contents. However, the degree of control seedlings was much more serious than that of transgenic lines and much more increases of the free proline contents in the transgenic lines than that in the control seedlings were observed. Meanwhile, lower extent decreases of the chlorophyll contents were detected in the transgenic seedlings. Quantitative RT-PCR was performed to analyze the expression of ten stress-related genes, including stress responsive transcription factor genes, ZmMYB59 and ZmMYC1, proline synthesis related genes, ZmP5CS1 and ZmP5CS2, photosynthesis-related genes, ZmELIP, ZmPSI-N, ZmOEE, Zmrbcs and ZmPLAS, and one ABA biosynthesis related gene, ZmSDR. The results showed that with the exception of ZmP5CS1 and ZmP5CS2 in line 9-10 and 19-11, ZmMYC1 in line 19-11 and ZmSDR in line 19-11, the expression of other stress-related genes were inhibited in transgenic lines under normal conditions. After salt treatment, the expressions of the ten stress-related genes were significantly induced in both wild-type (WT) and transgenic lines. However, compared to WT, the increases of ZmP5CS1 in all these three transgenic lines and ZmP5CS2 in line 9-10 were less than WT plants. This study provides an effective approach of maize genetic engineering for improved nutritive quality and salt tolerance.
Figures



Similar articles
-
The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants.Planta. 2017 Dec;246(6):1215-1231. doi: 10.1007/s00425-017-2766-9. Epub 2017 Aug 31. Planta. 2017. PMID: 28861611
-
Maize WRKY114 gene negatively regulates salt-stress tolerance in transgenic rice.Plant Cell Rep. 2020 Jan;39(1):135-148. doi: 10.1007/s00299-019-02481-3. Epub 2019 Oct 28. Plant Cell Rep. 2020. PMID: 31659429
-
Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance.Plant Biotechnol J. 2010 May 1;8(4):476-88. doi: 10.1111/j.1467-7652.2009.00492.x. Epub 2010 Mar 11. Plant Biotechnol J. 2010. PMID: 20233336
-
Overexpression of StDREB1 transcription factor increases tolerance to salt in transgenic potato plants.Mol Biotechnol. 2013 Jul;54(3):803-17. doi: 10.1007/s12033-012-9628-2. Mol Biotechnol. 2013. PMID: 23250722
-
Seed-specific expression of the lysine-rich protein gene sb401 significantly increases both lysine and total protein content in maize seeds.Food Nutr Bull. 2005 Dec;26(4):427-31. Food Nutr Bull. 2005. PMID: 16465991 Review.
Cited by
-
Filamentous cyanobacteria triples oil production in seawater-based medium supplemented with industrial waste: monosodium glutamate residue.Biotechnol Biofuels. 2019 Mar 14;12:53. doi: 10.1186/s13068-019-1391-1. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 30911333 Free PMC article.
-
Spatial and temporal activity of the foxtail millet (Setaria italica) seed-specific promoter pF128.Planta. 2015 Jan;241(1):57-67. doi: 10.1007/s00425-014-2164-5. Epub 2014 Sep 11. Planta. 2015. PMID: 25204632
-
Improving protein content and quality by over-expressing artificially synthetic fusion proteins with high lysine and threonine constituent in rice plants.Sci Rep. 2016 Sep 28;6:34427. doi: 10.1038/srep34427. Sci Rep. 2016. PMID: 27677708 Free PMC article.
-
Seed-Specific Expression of the Arabidopsis AtMAP18 Gene Increases both Lysine and Total Protein Content in Maize.PLoS One. 2015 Nov 18;10(11):e0142952. doi: 10.1371/journal.pone.0142952. eCollection 2015. PLoS One. 2015. PMID: 26580206 Free PMC article.
-
Physiological and Metabolic Responses of Gac Leaf (Momordica cochinchinensis (Lour.) Spreng.) to Salinity Stress.Plants (Basel). 2022 Sep 20;11(19):2447. doi: 10.3390/plants11192447. Plants (Basel). 2022. PMID: 36235312 Free PMC article.
References
-
- Osborne T.B., Mendel L.B. Nutritional properties of proteins of the maize kernel. J. Biol. Chem. 1914;18:1–16.
-
- Mertz E.T., Bates L.S., Nelson O.E. A mutant gene that changes the protein composition and increases the lysine content of maize endosperm. Science. 1964;145:279–280. - PubMed
-
- Vasal S.K., Villegas E., Bjarnason M., Gelaw B., Goertz P. Genetic Modifiers and Breeding Strategies in Developing Hard Endosperm opaque-2 Materials. In: Pollmer W.G., Phipps R.H., editors. Improvement of Quality Traits of Maize for Grain and Silage Use. 2nd ed. Martinus Nijhoff; London, UK: 1980. pp. 37–73.
-
- Huang S., Adams W.R., Zhou Q., Malloy K.P., Voyles D.A., Anthony J., Kriz A.L., Luethy M.H. Improving nutritional quality of maize proteins by expressing sense and antisense zein genes. J. Agric. Food Chem. 2004;52:1958–1964. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

