Deacetylation of p53 induces autophagy by suppressing Bmf expression
- PMID: 23629966
- PMCID: PMC3639396
- DOI: 10.1083/jcb.201205064
Deacetylation of p53 induces autophagy by suppressing Bmf expression
Abstract
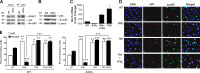
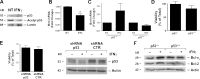
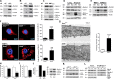
Interferon γ (IFN-γ)-induced cell death is mediated by the BH3-only domain protein, Bik, in a p53-independent manner. However, the effect of IFN-γ on p53 and how this affects autophagy have not been reported. The present study demonstrates that IFN-γ down-regulated expression of the BH3 domain-only protein, Bmf, in human and mouse airway epithelial cells in a p53-dependent manner. p53 also suppressed Bmf expression in response to other cell death-stimulating agents, including ultraviolet radiation and histone deacetylase inhibitors. IFN-γ did not affect Bmf messenger RNA half-life but increased nuclear p53 levels and the interaction of p53 with the Bmf promoter. IFN-γ-induced interaction of HDAC1 and p53 resulted in the deacetylation of p53 and suppression of Bmf expression independent of p53's proline-rich domain. Suppression of Bmf facilitated IFN-γ-induced autophagy by reducing the interaction of Beclin-1 and Bcl-2. Furthermore, autophagy was prominent in cultured bmf(-/-) but not in bmf(+/+) cells. Collectively, these observations show that deacetylation of p53 suppresses Bmf expression and facilitates autophagy.
Figures
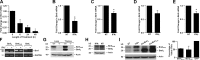



Comment in
-
BH3-only proteins, Bmf and Bim, in autophagy.Cell Cycle. 2013 Nov 15;12(22):3453-4. doi: 10.4161/cc.26696. Epub 2013 Oct 8. Cell Cycle. 2013. PMID: 24107625 Free PMC article. No abstract available.
References
-
- Coultas L., Bouillet P., Stanley E.G., Brodnicki T.C., Adams J.M., Strasser A. 2004. Proapoptotic BH3-only Bcl-2 family member Bik/Blk/Nbk is expressed in hemopoietic and endothelial cells but is redundant for their programmed death. Mol. Cell. Biol. 24:1570–1581 10.1128/MCB.24.4.1570-1581.2004 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

