Single-molecule DNA repair in live bacteria
- PMID: 23630273
- PMCID: PMC3657774
- DOI: 10.1073/pnas.1301804110
Single-molecule DNA repair in live bacteria
Abstract
Cellular DNA damage is reversed by balanced repair pathways that avoid accumulation of toxic intermediates. Despite their importance, the organization of DNA repair pathways and the function of repair enzymes in vivo have remained unclear because of the inability to directly observe individual reactions in living cells. Here, we used photoactivation, localization, and tracking in live Escherichia coli to directly visualize single fluorescent labeled DNA polymerase I (Pol) and ligase (Lig) molecules searching for DNA gaps and nicks, performing transient reactions, and releasing their products. Our general approach provides enzymatic rates and copy numbers, substrate-search times, diffusion characteristics, and the spatial distribution of reaction sites, at the single-cell level, all in one measurement. Single repair events last 2.1 s (Pol) and 2.5 s (Lig), respectively. Pol and Lig activities increased fivefold over the basal level within minutes of DNA methylation damage; their rates were limited by upstream base excision repair pathway steps. Pol and Lig spent >80% of their time searching for free substrates, thereby minimizing both the number and lifetime of toxic repair intermediates. We integrated these single-molecule observations to generate a quantitative, systems-level description of a model repair pathway in vivo.
Keywords: DNA damage response; cytosolic diffusion; protein-DNA interaction; single-molecule tracking; super-resolution microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

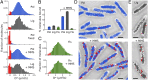
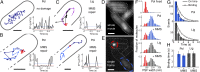


References
-
- Friedberg EC. DNA Repair and Mutagenesis. Washington, DC: American Society for Microbiology; 2006.
-
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

