Single-cell paired-end genome sequencing reveals structural variation per cell cycle
- PMID: 23630320
- PMCID: PMC3695511
- DOI: 10.1093/nar/gkt345
Single-cell paired-end genome sequencing reveals structural variation per cell cycle
Abstract
The nature and pace of genome mutation is largely unknown. Because standard methods sequence DNA from populations of cells, the genetic composition of individual cells is lost, de novo mutations in cells are concealed within the bulk signal and per cell cycle mutation rates and mechanisms remain elusive. Although single-cell genome analyses could resolve these problems, such analyses are error-prone because of whole-genome amplification (WGA) artefacts and are limited in the types of DNA mutation that can be discerned. We developed methods for paired-end sequence analysis of single-cell WGA products that enable (i) detecting multiple classes of DNA mutation, (ii) distinguishing DNA copy number changes from allelic WGA-amplification artefacts by the discovery of matching aberrantly mapping read pairs among the surfeit of paired-end WGA and mapping artefacts and (iii) delineating the break points and architecture of structural variants. By applying the methods, we capture DNA copy number changes acquired over one cell cycle in breast cancer cells and in blastomeres derived from a human zygote after in vitro fertilization. Furthermore, we were able to discover and fine-map a heritable inter-chromosomal rearrangement t(1;16)(p36;p12) by sequencing a single blastomere. The methods will expedite applications in basic genome research and provide a stepping stone to novel approaches for clinical genetic diagnosis.
Figures


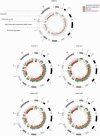
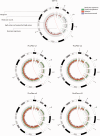
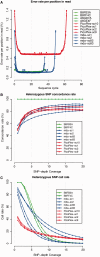
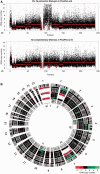
References
-
- Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–1558. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

