Integrating anatomy and function for zebrafish circuit analysis
- PMID: 23630469
- PMCID: PMC3632786
- DOI: 10.3389/fncir.2013.00074
Integrating anatomy and function for zebrafish circuit analysis
Abstract
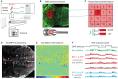
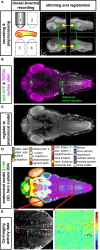
Due to its transparency, virtually every brain structure of the larval zebrafish is accessible to light-based interrogation of circuit function. Advanced stimulation techniques allow the activation of optogenetic actuators at different resolution levels, and genetically encoded calcium indicators report the activity of a large proportion of neurons in the CNS. Large datasets result and need to be analyzed to identify cells that have specific properties-e.g., activity correlation to sensory stimulation or behavior. Advances in three-dimensional (3D) functional mapping in zebrafish are promising; however, the mere coordinates of implicated neurons are not sufficient. To comprehensively understand circuit function, these functional maps need to be placed into the proper context of morphological features and projection patterns, neurotransmitter phenotypes, and key anatomical landmarks. We discuss the prospect of merging functional and anatomical data in an integrated atlas from the perspective of our work on long-range dopaminergic neuromodulation and the oculomotor system. We propose that such a resource would help researchers to surpass current hurdles in circuit analysis to achieve an integrated understanding of anatomy and function.
Keywords: brain anatomy; calcium imaging; neural circuits; neuronal types; optogenetics; zebrafish model system.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

