Improved antisense oligonucleotide design to suppress aberrant SMN2 gene transcript processing: towards a treatment for spinal muscular atrophy
- PMID: 23630626
- PMCID: PMC3632594
- DOI: 10.1371/journal.pone.0062114
Improved antisense oligonucleotide design to suppress aberrant SMN2 gene transcript processing: towards a treatment for spinal muscular atrophy
Abstract
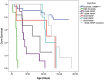
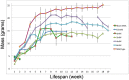
Spinal muscular atrophy (SMA) is caused by loss of the Survival Motor Neuron 1 (SMN1) gene, resulting in reduced SMN protein. Humans possess the additional SMN2 gene (or genes) that does produce low level of full length SMN, but cannot adequately compensate for loss of SMN1 due to aberrant splicing. The majority of SMN2 gene transcripts lack exon 7 and the resultant SMNΔ7 mRNA is translated into an unstable and non-functional protein. Splice intervention therapies to promote exon 7 retention and increase amounts of full-length SMN2 transcript offer great potential as a treatment for SMA patients. Several splice silencing motifs in SMN2 have been identified as potential targets for antisense oligonucleotide mediated splice modification. A strong splice silencer is located downstream of exon 7 in SMN2 intron 7. Antisense oligonucleotides targeting this motif promoted SMN2 exon 7 retention in the mature SMN2 transcripts, with increased SMN expression detected in SMA fibroblasts. We report here systematic optimisation of phosphorodiamidate morpholino oligonucleotides (PMO) that promote exon 7 retention to levels that rescued the phenotype in a severe mouse model of SMA after intracerebroventricular delivery. Furthermore, the PMO gives the longest survival reported to date after a single dosing by ICV.
Conflict of interest statement
Figures



Similar articles
-
A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse.Hum Mol Genet. 2012 Apr 1;21(7):1625-38. doi: 10.1093/hmg/ddr600. Epub 2011 Dec 20. Hum Mol Genet. 2012. PMID: 22186025 Free PMC article.
-
Combination of valproic acid and morpholino splice-switching oligonucleotide produces improved outcomes in spinal muscular atrophy patient-derived fibroblasts.Neurochem Int. 2017 Sep;108:213-221. doi: 10.1016/j.neuint.2017.02.016. Epub 2017 Apr 4. Neurochem Int. 2017. PMID: 28389270
-
Antisense Oligonucleotide Induction of the hnRNPA1b Isoform Affects Pre-mRNA Splicing of SMN2 in SMA Type I Fibroblasts.Int J Mol Sci. 2022 Apr 1;23(7):3937. doi: 10.3390/ijms23073937. Int J Mol Sci. 2022. PMID: 35409296 Free PMC article.
-
Recent Advances and Clinical Applications of Exon Inclusion for Spinal Muscular Atrophy.Methods Mol Biol. 2018;1828:57-68. doi: 10.1007/978-1-4939-8651-4_3. Methods Mol Biol. 2018. PMID: 30171534 Review.
-
Antisense oligonucleotides for the treatment of spinal muscular atrophy.Hum Gene Ther. 2013 May;24(5):489-98. doi: 10.1089/hum.2012.225. Hum Gene Ther. 2013. PMID: 23544870 Free PMC article. Review.
Cited by
-
Human RNAi pathway: crosstalk with organelles and cells.Funct Integr Genomics. 2014 Mar;14(1):31-46. doi: 10.1007/s10142-013-0344-1. Epub 2013 Nov 7. Funct Integr Genomics. 2014. PMID: 24197738 Review.
-
Spinal muscular atrophy: development and implementation of potential treatments.Ann Neurol. 2013 Sep;74(3):348-62. doi: 10.1002/ana.23995. Ann Neurol. 2013. PMID: 23939659 Free PMC article. Review.
-
Optimization of Morpholino Antisense Oligonucleotides Targeting the Intronic Repressor Element1 in Spinal Muscular Atrophy.Mol Ther. 2016 Sep;24(9):1592-601. doi: 10.1038/mt.2016.145. Epub 2016 Jul 9. Mol Ther. 2016. PMID: 27401142 Free PMC article.
-
Survival Motor Neuron (SMN) protein is required for normal mouse liver development.Sci Rep. 2016 Oct 4;6:34635. doi: 10.1038/srep34635. Sci Rep. 2016. PMID: 27698380 Free PMC article.
-
Potential of Cell-Penetrating Peptide-Conjugated Antisense Oligonucleotides for the Treatment of SMA.Molecules. 2024 Jun 4;29(11):2658. doi: 10.3390/molecules29112658. Molecules. 2024. PMID: 38893532 Free PMC article. Review.
References
-
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, et al. (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80: 155–165. - PubMed
-
- Gennarelli M, Lucarelli M, Capon F, Pizzuti A, Merlini L, et al. (1995) Survival motor neuron gene transcript analysis in muscles from spinal muscular atrophy patients. Biochemical and biophysical research communications 213: 342–348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

