High resolution two-dimensional electrophoresis of proteins
- PMID: 236308
- PMCID: PMC2874754
High resolution two-dimensional electrophoresis of proteins
Abstract
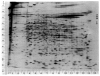
A technique has been developed for the separation of proteins by two-dimensional polyacrylamide gel electrophoresis. Due to its resolution and sensitivity, this technique is a powerful tool for the analysis and detection of proteins from complex biological sources. Proteins are separated according to isoelectric point by isoelectric focusing in the first dimension, and according to molecular weight by sodium dodecyl sulfate electrophoresis in the second dimension. Since these two parameters are unrelated, it is possible to obtain an almost uniform distribution of protein spots across a two-diminsional gel. This technique has resolved 1100 different components from Escherichia coli and should be capable of resolving a maximum of 5000 proteins. A protein containing as little as one disintegration per min of either 14C or 35S can be detected by autoradiography. A protein which constitutes 10 minus 4 to 10 minus 5% of the total protein can be detected and quantified by autoradiography. The reproducibility of the separation is sufficient to permit each spot on one separation to be matched with a spot on a different separation. This technique provides a method for estimation (at the described sensitivities) of the number of proteins made by any biological system. This system can resolve proteins differing in a single charge and consequently can be used in the analysis of in vivo modifications resulting in a change in charge. Proteins whose charge is changed by missense mutations can be identified. A detailed description of the methods as well as the characteristics of this system are presented.
Figures
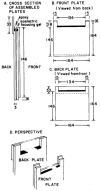


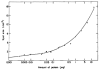

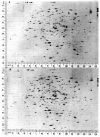
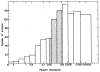
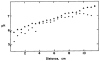
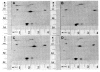

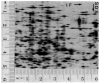
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

