Structural diversity of eukaryotic membrane cytochrome p450s
- PMID: 23632020
- PMCID: PMC3682514
- DOI: 10.1074/jbc.R113.452805
Structural diversity of eukaryotic membrane cytochrome p450s
Abstract
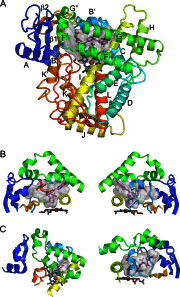
X-ray crystal structures are available for 29 eukaryotic microsomal, chloroplast, or mitochondrial cytochrome P450s, including two non-monooxygenase P450s. These structures provide a basis for understanding structure-function relations that underlie their distinct catalytic activities. Moreover, structural plasticity has been characterized for individual P450s that aids in understanding substrate binding in P450s that mediate drug clearance.
Keywords: Carcinogenesis; Cytochrome P450; Drug Development; Drug Metabolism; Protein structure; Steroidogenesis.
Figures

References
-
- Nelson D. R., Zeldin D. C., Hoffman S. M., Maltais L. J., Wain H. M., Nebert D. W. (2004) Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics 14, 1–18 - PubMed
-
- Williams P. A., Cosme J., Sridhar V., Johnson E. F., McRee D. E. (2000) The crystallographic structure of a mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol. Cell 5, 121–131 - PubMed
-
- Poulos T. L., Finzel B. C., Gunsalus I. C., Wagner G. C., Kraut J. (1985) The 2.6-Å crystal structure of the Pseudomonas putida cytochrome P-450. J. Biol. Chem. 260, 16122–16130 - PubMed
-
- Poulos T. L., Johnson E. F. (2005) Structures of cytochrome P450 enzymes. In Cytochrome P450: Structure, Mechanism, and Biochemistry (Ortiz de Montellano P. R., ed) 3rd Ed., pp. 87–114, Kluwer Academic/Plenum Publishers, New York
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

