Genome conformation capture reveals that the Escherichia coli chromosome is organized by replication and transcription
- PMID: 23632166
- PMCID: PMC3695519
- DOI: 10.1093/nar/gkt325
Genome conformation capture reveals that the Escherichia coli chromosome is organized by replication and transcription
Abstract
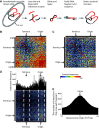
To fit within the confines of the cell, bacterial chromosomes are highly condensed into a structure called the nucleoid. Despite the high degree of compaction in the nucleoid, the genome remains accessible to essential biological processes, such as replication and transcription. Here, we present the first high-resolution chromosome conformation capture-based molecular analysis of the spatial organization of the Escherichia coli nucleoid during rapid growth in rich medium and following an induced amino acid starvation that promotes the stringent response. Our analyses identify the presence of origin and terminus domains in exponentially growing cells. Moreover, we observe an increased number of interactions within the origin domain and significant clustering of SeqA-binding sequences, suggesting a role for SeqA in clustering of newly replicated chromosomes. By contrast, 'histone-like' protein (i.e. Fis, IHF and H-NS) -binding sites did not cluster, and their role in global nucleoid organization does not manifest through the mediation of chromosomal contacts. Finally, genes that were downregulated after induction of the stringent response were spatially clustered, indicating that transcription in E. coli occurs at transcription foci.
Figures




References
-
- Woldringh CL, Nanninga N. Structural and physical aspects of bacterial chromosome segregation. J. Struct. Biol. 2006;156:273–283. - PubMed
-
- Ishihama A. The nucleoid: an overview. In: Curtiss R, Kaper JB, Squires CL, Karp PD, Neidhardt FC, Slauch JM, editors. EcoSal. Washington, DC: ASM Press; 2009.
-
- Sherratt DJ. Bacterial chromosome dynamics. Science. 2003;301:780–785. - PubMed
-
- Mercier R, Petit MA, Schbath S, Robin S, El Karoui M, Boccard F, Espeli O. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell. 2008;135:475–485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

