Conventional and non-conventional Drosophila Toll signaling
- PMID: 23632253
- PMCID: PMC3787077
- DOI: 10.1016/j.dci.2013.04.011
Conventional and non-conventional Drosophila Toll signaling
Abstract
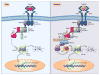
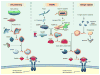
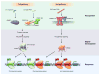
The discovery of Toll in Drosophila and of the remarkable conservation in pathway composition and organization catalyzed a transformation in our understanding of innate immune recognition and response. At the center of that picture is a cascade of interactions in which specific microbial cues activate Toll receptors, which then transmit signals driving transcription factor nuclear localization and activity. Experiments gave substance to the vision of pattern recognition receptors, linked phenomena in development, gene regulation, and immunity into a coherent whole, and revealed a rich set of variations for identifying non-self and responding effectively. More recently, research in Drosophila has illuminated the positive and negative regulation of Toll activation, the organization of signaling events at and beneath membranes, the sorting of information flow, and the existence of non-conventional signaling via Toll-related receptors. Here, we provide an overview of the Toll pathway of flies and highlight these ongoing realms of research.
Keywords: Drosophila; Innate immunity; NF-κB; Non-conventional pathway; Toll.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
References
-
- Anderson KV, Nusslein VC. Information for the dorsal-ventral pattern of the Drosophila embryo is stored as maternal mRNA. Nature. 1984;311:223–227. - PubMed
-
- Belvin MP, Anderson KV. A conserved signaling pathway: the Drosophila toll-dorsal pathway. Ann Rev Cell Dev Biol. 1996;12:393–416. - PubMed
-
- Bilak H, Tauszig-Delamasure S, Imler JL. Toll and Toll-like receptors in Drosophila. Biochem Soc Trans. 2003;31:648–651. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

