Thermostable DNA ligase-mediated PCR production of circular plasmid (PPCP) and its application in directed evolution via in situ error-prone PCR
- PMID: 23633530
- PMCID: PMC3738163
- DOI: 10.1093/dnares/dst016
Thermostable DNA ligase-mediated PCR production of circular plasmid (PPCP) and its application in directed evolution via in situ error-prone PCR
Abstract
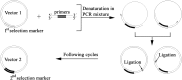
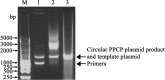
Polymerase chain reaction (PCR) is a powerful method to produce linear DNA fragments. Here we describe the Tma thermostable DNA ligase-mediated PCR production of circular plasmid (PPCP) and its application in directed evolution via in situ error-prone PCR. In this thermostable DNA ligase-mediated whole-plasmid amplification method, the resultant DNA nick between the 5' end of the PCR primer and the extended newly synthesized DNA 3' end of each PCR cycle is ligated by Tma DNA ligase, resulting in circular plasmid DNA product that can be directly transformed. The template plasmid DNA is eliminated by 'selection marker swapping' upon transformation. When performed under an error-prone condition with Taq DNA polymerase, PPCP allows one-step construction of mutagenesis libraries based on in situ error-prone PCR so that random mutations are introduced into the target gene without altering the expression vector plasmid. A significant difference between PPCP and previously published methods is that PPCP allows exponential amplification of circular DNA. We used this method to create random mutagenesis libraries of a xylanase gene and two cellulase genes. Screening of these libraries resulted in mutant proteins with desired properties, demonstrating the usefulness of in situ error-prone PPCP for creating random mutagenesis libraries for directed evolution.
Keywords: amplification of circular plasmids; directed evolution; error-prone PCR; random mutagenesis libraries; thermostable DNA ligase.
Figures

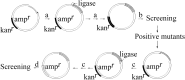

Similar articles
-
Development and Use of a Novel Random Mutagenesis Method: In Situ Error-Prone PCR (is-epPCR).Methods Mol Biol. 2017;1498:497-506. doi: 10.1007/978-1-4939-6472-7_34. Methods Mol Biol. 2017. PMID: 27709598
-
Polishing the craft of genetic diversity creation in directed evolution.Biotechnol Adv. 2013 Dec;31(8):1707-21. doi: 10.1016/j.biotechadv.2013.08.021. Epub 2013 Sep 6. Biotechnol Adv. 2013. PMID: 24012599 Review.
-
Error-prone PCR of a fungal xylanase for improvement of its alkaline and thermal stability.FEMS Microbiol Lett. 2009 Apr;293(1):42-7. doi: 10.1111/j.1574-6968.2009.01519.x. Epub 2009 Feb 7. FEMS Microbiol Lett. 2009. PMID: 19220468
-
Restriction enzyme-free construction of random gene mutagenesis libraries in Escherichia coli.Anal Biochem. 2012 Feb 15;421(2):640-8. doi: 10.1016/j.ab.2011.11.009. Epub 2011 Nov 18. Anal Biochem. 2012. PMID: 22155067 Free PMC article.
-
Mutant library construction in directed molecular evolution: casting a wider net.Mol Biotechnol. 2006 Sep;34(1):55-68. doi: 10.1385/MB:34:1:55. Mol Biotechnol. 2006. PMID: 16943572 Review.
Cited by
-
Thermostable DNA ligases from hyperthermophiles in biotechnology.Front Microbiol. 2023 May 24;14:1198784. doi: 10.3389/fmicb.2023.1198784. eCollection 2023. Front Microbiol. 2023. PMID: 37293226 Free PMC article. Review.
-
Self-assembly of a multimeric genomic hydrogel via multi-primed chain reaction of dual single-stranded circular plasmids for cell-free protein production.iScience. 2023 Jun 10;26(7):107089. doi: 10.1016/j.isci.2023.107089. eCollection 2023 Jul 21. iScience. 2023. PMID: 37416467 Free PMC article.
-
PCR performance of a thermostable heterodimeric archaeal DNA polymerase.Front Microbiol. 2014 May 7;5:195. doi: 10.3389/fmicb.2014.00195. eCollection 2014. Front Microbiol. 2014. PMID: 24847315 Free PMC article.
References
-
- Chirumamilla R.R., Muralidhar R., Marchant R., Nigam P. Improving the quality of industrially important enzymes by directed evolution. Mol. Cell Biochem. 2001;224:159–68. doi:10.1023/A:1011904405002. - DOI - PubMed
-
- Cherry J.R., Fidantsef A.L. Directed evolution of industrial enzymes: an update. Curr. Opin. Biotechnol. 2003;14:438–43. doi:10.1016/S0958-1669(03)00099-5. - DOI - PubMed
-
- Eijsink V.G.H., Gaseidnes S., Borchert T.V., van den Burg B. Directed evolution of enzyme stability. Biomol. Eng. 2005;22:21–30. doi:10.1016/j.bioeng.2004.12.003. - DOI - PubMed
-
- Yuan L., Kurek I., English J., Keenan R. Laboratory-directed protein evolution. Microbiol. Mol. Biol. Rev. 2005;69:373–92. doi:10.1128/MMBR.69.3.373-392.2005. - DOI - PMC - PubMed
-
- Otten L.G., Quax W.J. Directed evolution: selecting today's biocatalysts. Biomol. Eng. 2005;22:1–9. doi:10.1016/j.bioeng.2005.02.002. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

