AutoDrug: fully automated macromolecular crystallography workflows for fragment-based drug discovery
- PMID: 23633588
- PMCID: PMC3640469
- DOI: 10.1107/S0907444913001984
AutoDrug: fully automated macromolecular crystallography workflows for fragment-based drug discovery
Abstract
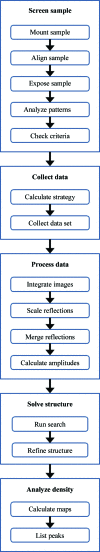
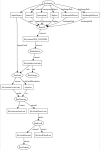
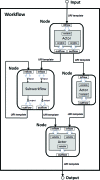
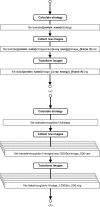
AutoDrug is software based upon the scientific workflow paradigm that integrates the Stanford Synchrotron Radiation Lightsource macromolecular crystallography beamlines and third-party processing software to automate the crystallography steps of the fragment-based drug-discovery process. AutoDrug screens a cassette of fragment-soaked crystals, selects crystals for data collection based on screening results and user-specified criteria and determines optimal data-collection strategies. It then collects and processes diffraction data, performs molecular replacement using provided models and detects electron density that is likely to arise from bound fragments. All processes are fully automated, i.e. are performed without user interaction or supervision. Samples can be screened in groups corresponding to particular proteins, crystal forms and/or soaking conditions. A single AutoDrug run is only limited by the capacity of the sample-storage dewar at the beamline: currently 288 samples. AutoDrug was developed in conjunction with RestFlow, a new scientific workflow-automation framework. RestFlow simplifies the design of AutoDrug by managing the flow of data and the organization of results and by orchestrating the execution of computational pipeline steps. It also simplifies the execution and interaction of third-party programs and the beamline-control system. Modeling AutoDrug as a scientific workflow enables multiple variants that meet the requirements of different user groups to be developed and supported. A workflow tailored to mimic the crystallography stages comprising the drug-discovery pipeline of CoCrystal Discovery Inc. has been deployed and successfully demonstrated. This workflow was run once on the same 96 samples that the group had examined manually and the workflow cycled successfully through all of the samples, collected data from the same samples that were selected manually and located the same peaks of unmodeled density in the resulting difference Fourier maps.
Keywords: AutoDrug; fragment-based drug discovery; workflow automation.
Figures
References
-
- Bavoil, L., Callahan, S., Crossno, P., Freire, J., Scheidegger, C., Silva, C. & Vo, H. (2005). Visualization, 2005. VIS 05. IEEE, pp. 135–142. doi:10.1109/VISUAL.2005.1532788.
-
- Bourenkov, G. P. & Popov, A. N. (2006). Acta Cryst. D62, 58–64. - PubMed
-
- Bowers, S., Ludaescher, B., Ngu, A. H. & Critchlow, T. (2006). Workshop on Workflow and Data Flow for Scientific Applications (SciFlow)
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

