A refined model of the prototypical Salmonella SPI-1 T3SS basal body reveals the molecular basis for its assembly
- PMID: 23633951
- PMCID: PMC3635987
- DOI: 10.1371/journal.ppat.1003307
A refined model of the prototypical Salmonella SPI-1 T3SS basal body reveals the molecular basis for its assembly
Abstract
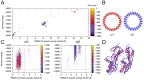
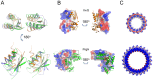
The T3SS injectisome is a syringe-shaped macromolecular assembly found in pathogenic Gram-negative bacteria that allows for the direct delivery of virulence effectors into host cells. It is composed of a "basal body", a lock-nut structure spanning both bacterial membranes, and a "needle" that protrudes away from the bacterial surface. A hollow channel spans throughout the apparatus, permitting the translocation of effector proteins from the bacterial cytosol to the host plasma membrane. The basal body is composed largely of three membrane-embedded proteins that form oligomerized concentric rings. Here, we report the crystal structures of three domains of the prototypical Salmonella SPI-1 basal body, and use a new approach incorporating symmetric flexible backbone docking and EM data to produce a model for their oligomeric assembly. The obtained models, validated by biochemical and in vivo assays, reveal the molecular details of the interactions driving basal body assembly, and notably demonstrate a conserved oligomerization mechanism.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Worrall LJ, Lameignere E, Strynadka NC (2011) Structural overview of the bacterial injectisome. Curr Opin Microbiol 14: 3–8. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

