High-resolution mapping of h1 linker histone variants in embryonic stem cells
- PMID: 23633960
- PMCID: PMC3636266
- DOI: 10.1371/journal.pgen.1003417
High-resolution mapping of h1 linker histone variants in embryonic stem cells
Abstract
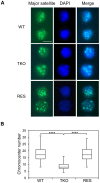
H1 linker histones facilitate higher-order chromatin folding and are essential for mammalian development. To achieve high-resolution mapping of H1 variants H1d and H1c in embryonic stem cells (ESCs), we have established a knock-in system and shown that the N-terminally tagged H1 proteins are functionally interchangeable to their endogenous counterparts in vivo. H1d and H1c are depleted from GC- and gene-rich regions and active promoters, inversely correlated with H3K4me3, but positively correlated with H3K9me3 and associated with characteristic sequence features. Surprisingly, both H1d and H1c are significantly enriched at major satellites, which display increased nucleosome spacing compared with bulk chromatin. While also depleted at active promoters and enriched at major satellites, overexpressed H1(0) displays differential binding patterns in specific repetitive sequences compared with H1d and H1c. Depletion of H1c, H1d, and H1e causes pericentric chromocenter clustering and de-repression of major satellites. These results integrate the localization of an understudied type of chromatin proteins, namely the H1 variants, into the epigenome map of mouse ESCs, and we identify significant changes at pericentric heterochromatin upon depletion of this epigenetic mark.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Reduction of Hox gene expression by histone H1 depletion.PLoS One. 2012;7(6):e38829. doi: 10.1371/journal.pone.0038829. Epub 2012 Jun 11. PLoS One. 2012. PMID: 22701719 Free PMC article.
-
Developmentally regulated linker histone H1c promotes heterochromatin condensation and mediates structural integrity of rod photoreceptors in mouse retina.J Biol Chem. 2013 Jun 14;288(24):17895-907. doi: 10.1074/jbc.M113.452144. Epub 2013 May 3. J Biol Chem. 2013. PMID: 23645681 Free PMC article.
-
Histone h1 depletion impairs embryonic stem cell differentiation.PLoS Genet. 2012;8(5):e1002691. doi: 10.1371/journal.pgen.1002691. Epub 2012 May 10. PLoS Genet. 2012. PMID: 22589736 Free PMC article.
-
The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle.EMBO Rep. 2015 Nov;16(11):1439-53. doi: 10.15252/embr.201540749. Epub 2015 Oct 15. EMBO Rep. 2015. PMID: 26474902 Free PMC article. Review.
-
Regulation of chromatin structure by histone H3S10 phosphorylation.Chromosome Res. 2006;14(4):393-404. doi: 10.1007/s10577-006-1063-4. Chromosome Res. 2006. PMID: 16821135 Review.
Cited by
-
Identification of lamin B-regulated chromatin regions based on chromatin landscapes.Mol Biol Cell. 2015 Jul 15;26(14):2685-97. doi: 10.1091/mbc.E15-04-0210. Epub 2015 May 20. Mol Biol Cell. 2015. PMID: 25995381 Free PMC article.
-
H1 linker histones silence repetitive elements by promoting both histone H3K9 methylation and chromatin compaction.Proc Natl Acad Sci U S A. 2020 Jun 23;117(25):14251-14258. doi: 10.1073/pnas.1920725117. Epub 2020 Jun 8. Proc Natl Acad Sci U S A. 2020. PMID: 32513732 Free PMC article.
-
Histone h1.3 suppresses h19 noncoding RNA expression and cell growth of ovarian cancer cells.Cancer Res. 2014 Nov 15;74(22):6463-73. doi: 10.1158/0008-5472.CAN-13-2922. Epub 2014 Sep 9. Cancer Res. 2014. PMID: 25205099 Free PMC article.
-
Histone H1 Mutations in Lymphoma: A Link(er) between Chromatin Organization, Developmental Reprogramming, and Cancer.Cancer Res. 2021 Dec 15;81(24):6061-6070. doi: 10.1158/0008-5472.CAN-21-2619. Epub 2021 Sep 27. Cancer Res. 2021. PMID: 34580064 Free PMC article. Review.
-
H1 histones control the epigenetic landscape by local chromatin compaction.Nature. 2021 Jan;589(7841):293-298. doi: 10.1038/s41586-020-3032-z. Epub 2020 Dec 9. Nature. 2021. PMID: 33299182 Free PMC article.
References
-
- Wolffe AP (1998) Chromatin: Structure and Function. San Diego, CA: Academic Press.
-
- van Holde KE (1989) Chromatin: New York: Springer-Verlag.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

