Characterization of sex determination and sex differentiation genes in Latimeria
- PMID: 23634199
- PMCID: PMC3636272
- DOI: 10.1371/journal.pone.0056006
Characterization of sex determination and sex differentiation genes in Latimeria
Abstract
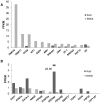
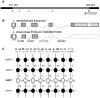
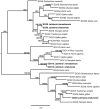
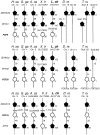
Genes involved in sex determination and differentiation have been identified in mice, humans, chickens, reptiles, amphibians and teleost fishes. However, little is known of their functional conservation, and it is unclear whether there is a common set of genes shared by all vertebrates. Coelacanths, basal Sarcopterygians and unique "living fossils", could help establish an inventory of the ancestral genes involved in these important developmental processes and provide insights into their components. In this study 33 genes from the genome of Latimeria chalumnae and from the liver and testis transcriptomes of Latimeria menadoensis, implicated in sex determination and differentiation, were identified and characterized and their expression levels measured. Interesting findings were obtained for GSDF, previously identified only in teleosts and now characterized for the first time in the sarcopterygian lineage; FGF9, which is not found in teleosts; and DMRT1, whose expression in adult gonads has recently been related to maintenance of sexual identity. The gene repertoire and testis-specific gene expression documented in coelacanths demonstrate a greater similarity to modern fishes and point to unexpected changes in the gene regulatory network governing sexual development.
Conflict of interest statement
Figures




Similar articles
-
Transcriptome display during tilapia sex determination and differentiation as revealed by RNA-Seq analysis.BMC Genomics. 2018 May 15;19(1):363. doi: 10.1186/s12864-018-4756-0. BMC Genomics. 2018. PMID: 29764377 Free PMC article.
-
A Comparative View on Sex Differentiation and Gametogenesis Genes in Lungfish and Coelacanths.Genome Biol Evol. 2018 Jun 1;10(6):1430-1444. doi: 10.1093/gbe/evy101. Genome Biol Evol. 2018. PMID: 29850809 Free PMC article.
-
Involvement of Transforming Growth Factor Beta Family Genes in Gonadal Differentiation in Japanese Eel, Anguilla japonica, According to Sex-Related Gene Expressions.Cells. 2021 Nov 4;10(11):3007. doi: 10.3390/cells10113007. Cells. 2021. PMID: 34831230 Free PMC article.
-
Sex determination and gonadal sex differentiation in the chicken model.Int J Dev Biol. 2018;62(1-2-3):153-166. doi: 10.1387/ijdb.170319cs. Int J Dev Biol. 2018. PMID: 29616724 Review.
-
Insights of sex determination and differentiation from medaka as a teleost model.Yi Chuan. 2017 Jun 20;39(6):441-454. doi: 10.16288/j.yczz.17-140. Yi Chuan. 2017. PMID: 28903904 Review.
Cited by
-
Transcriptomics of two evolutionary novelties: how to make a sperm-transfer organ out of an anal fin and a sexually selected "sword" out of a caudal fin.Ecol Evol. 2015 Feb;5(4):848-64. doi: 10.1002/ece3.1390. Epub 2015 Jan 23. Ecol Evol. 2015. PMID: 25750712 Free PMC article.
-
Gene expression analysis at the onset of sex differentiation in turbot (Scophthalmus maximus).BMC Genomics. 2015 Nov 18;16:973. doi: 10.1186/s12864-015-2142-8. BMC Genomics. 2015. PMID: 26581195 Free PMC article.
-
Complete depletion of primordial germ cells in an All-female fish leads to Sex-biased gene expression alteration and sterile All-male occurrence.BMC Genomics. 2015 Nov 18;16:971. doi: 10.1186/s12864-015-2130-z. BMC Genomics. 2015. PMID: 26582363 Free PMC article.
-
Gonadal soma controls ovarian follicle proliferation through Gsdf in zebrafish.Dev Dyn. 2017 Nov;246(11):925-945. doi: 10.1002/dvdy.24579. Epub 2017 Sep 25. Dev Dyn. 2017. PMID: 28856758 Free PMC article.
-
De novo transcriptome based on next-generation sequencing reveals candidate genes with sex-specific expression in Arapaima gigas (Schinz, 1822), an ancient Amazonian freshwater fish.PLoS One. 2018 Oct 29;13(10):e0206379. doi: 10.1371/journal.pone.0206379. eCollection 2018. PLoS One. 2018. PMID: 30372461 Free PMC article.
References
-
- Hayes TB (1998) Sex determination and primary sex differentiation in amphibian: genetic and developmental mechanisms. J Exp Zool 281: 373–399. - PubMed
-
- Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, et al. (2009) Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139: 1130–1142. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

