Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia
- PMID: 23634996
- PMCID: PMC3767041
- DOI: 10.1056/NEJMoa1301689
Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia
Erratum in
- N Engl J Med. 2013 Jul 4;369(1):98
Abstract
Background: Many mutations that contribute to the pathogenesis of acute myeloid leukemia (AML) are undefined. The relationships between patterns of mutations and epigenetic phenotypes are not yet clear.
Methods: We analyzed the genomes of 200 clinically annotated adult cases of de novo AML, using either whole-genome sequencing (50 cases) or whole-exome sequencing (150 cases), along with RNA and microRNA sequencing and DNA-methylation analysis.
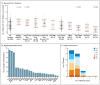
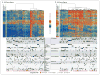
Results: AML genomes have fewer mutations than most other adult cancers, with an average of only 13 mutations found in genes. Of these, an average of 5 are in genes that are recurrently mutated in AML. A total of 23 genes were significantly mutated, and another 237 were mutated in two or more samples. Nearly all samples had at least 1 nonsynonymous mutation in one of nine categories of genes that are almost certainly relevant for pathogenesis, including transcription-factor fusions (18% of cases), the gene encoding nucleophosmin (NPM1) (27%), tumor-suppressor genes (16%), DNA-methylation-related genes (44%), signaling genes (59%), chromatin-modifying genes (30%), myeloid transcription-factor genes (22%), cohesin-complex genes (13%), and spliceosome-complex genes (14%). Patterns of cooperation and mutual exclusivity suggested strong biologic relationships among several of the genes and categories.
Conclusions: We identified at least one potential driver mutation in nearly all AML samples and found that a complex interplay of genetic events contributes to AML pathogenesis in individual patients. The databases from this study are widely available to serve as a foundation for further investigations of AML pathogenesis, classification, and risk stratification. (Funded by the National Institutes of Health.).
Figures



Comment in
-
The beginning of the end of the beginning in cancer genomics.N Engl J Med. 2013 May 30;368(22):2138-40. doi: 10.1056/NEJMe1303816. Epub 2013 May 1. N Engl J Med. 2013. PMID: 23634995 No abstract available.
-
Genetics: the AML mutational landscape.Nat Rev Clin Oncol. 2013 Jun;10(6):305. doi: 10.1038/nrclinonc.2013.81. Epub 2013 May 14. Nat Rev Clin Oncol. 2013. PMID: 23670658 No abstract available.
-
A panoramic view of acute myeloid leukemia.Nat Genet. 2013 Jun;45(6):586-7. doi: 10.1038/ng.2651. Nat Genet. 2013. PMID: 23715324
-
Cancer: Mutations close in on gene regulation.Nature. 2013 Jul 4;499(7456):35-6. doi: 10.1038/499035a. Nature. 2013. PMID: 23823789 No abstract available.
-
Genomic landscapes and clonality of de novo AML.N Engl J Med. 2013 Oct 10;369(15):1473. doi: 10.1056/NEJMc1308782. N Engl J Med. 2013. PMID: 24106950 Free PMC article. No abstract available.
-
Genomic landscapes and clonality of de novo AML.N Engl J Med. 2013 Oct 10;369(15):1472-3. doi: 10.1056/NEJMc1308782. N Engl J Med. 2013. PMID: 24106951 No abstract available.
-
Genetic and epigenetic map of acute myeloid leukemia.Pharmacogenomics. 2013 Dec;14(16):1950. Pharmacogenomics. 2013. PMID: 24422220 No abstract available.
References
-
- Rowley JD. Chromosomal translocations: revisited yet again. Blood. 2008;112:2183–9. - PubMed
-
- Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–36. - PubMed
-
- Bullinger L, Krönke J, Schön C, et al. Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia. 2010;24:438–49. - PubMed
-
- Suela J, Alvarez S, Cigudosa JC. DNA profiling by arrayCGH in acute myeloid leukemia and myelodysplastic syndromes. Cytogenet Genome Res. 2007;118:304–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U24CA143840/CA/NCI NIH HHS/United States
- P01CA101937/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- U24 CA143882/CA/NCI NIH HHS/United States
- U24 CA143866/CA/NCI NIH HHS/United States
- U24CA144025/CA/NCI NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- R01 HG005690/HG/NHGRI NIH HHS/United States
- U24 CA143843/CA/NCI NIH HHS/United States
- U24CA143867/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- U24CA143799/CA/NCI NIH HHS/United States
- U24 CA143883/CA/NCI NIH HHS/United States
- R01 CA162086/CA/NCI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U54HG003273/HG/NHGRI NIH HHS/United States
- U24 CA143835/CA/NCI NIH HHS/United States
- U24CA143858/CA/NCI NIH HHS/United States
- U24CA143882/CA/NCI NIH HHS/United States
- U54HG003067/HG/NHGRI NIH HHS/United States
- U24 CA143845/CA/NCI NIH HHS/United States
- U24 CA143799/CA/NCI NIH HHS/United States
- U24 CA144025/CA/NCI NIH HHS/United States
- U24CA143883/CA/NCI NIH HHS/United States
- U54HG003079/HG/NHGRI NIH HHS/United States
- U24 CA143840/CA/NCI NIH HHS/United States
- U24CA143835/CA/NCI NIH HHS/United States
- U24 CA143858/CA/NCI NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- U24CA143843/CA/NCI NIH HHS/United States
- U24 CA143848/CA/NCI NIH HHS/United States
- P01 CA101937/CA/NCI NIH HHS/United States
- U24CA143848/CA/NCI NIH HHS/United States
- U24CA143866/CA/NCI NIH HHS/United States
- U24CA143845/CA/NCI NIH HHS/United States
- R01 CA083962/CA/NCI NIH HHS/United States
- U24 CA143867/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
