Software for selecting the most informative sets of genomic loci for multi-target microbial typing
- PMID: 23635100
- PMCID: PMC3660239
- DOI: 10.1186/1471-2105-14-148
Software for selecting the most informative sets of genomic loci for multi-target microbial typing
Abstract
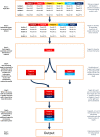
Background: High-throughput sequencing can identify numerous potential genomic targets for microbial strain typing, but identification of the most informative combinations requires the use of computational screening tools. This paper describes novel software-- Automated Selection of Typing Target Subsets (AuSeTTS)--that allows intelligent selection of optimal targets for pathogen strain typing. The objective of this software is to maximise both discriminatory power, using Simpson's index of diversity (D), and concordance with existing typing methods, using the adjusted Wallace coefficient (AW). The program interrogates molecular typing results for panels of isolates, based on large target sets, and iteratively examines each target, one-by-one, to determine the most informative subset.
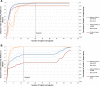
Results: AuSeTTS was evaluated using three target sets: 51 binary targets (13 toxin genes, 16 phage-related loci and 22 SCCmec elements), used for multilocus typing of 153 methicillin-resistant Staphylococcus aureus (MRSA) isolates; 17 MLVA loci in 502 Streptococcus pneumoniae isolates from the MLVA database (http://www.mlva.eu) and 12 MLST loci for 98 Cryptococcus spp. isolates.The maximum D for MRSA, 0.984, was achieved with a subset of 20 targets and a D value of 0.954 with 7 targets. Twelve targets predicted MLST with a maximum AW of 0.9994. All 17 S. pneumoniae MLVA targets were required to achieve maximum D of 0.997, but 4 targets reached D of 0.990. Twelve targets predicted pneumococcal serotype with a maximum AW of 0.899 and 9 predicted MLST with maximum AW of 0.963. Eight of the 12 MLST loci were sufficient to achieve the maximum D of 0.963 for Cryptococcus spp.
Conclusions: Computerised analysis with AuSeTTS allows rapid selection of the most discriminatory targets for incorporation into typing schemes. Output of the program is presented in both tabular and graphical formats and the software is available for free download from http://www.cidmpublichealth.org/pages/ausetts.html.
Figures
Similar articles
-
Multiple-locus variable number tandem repeat analysis for Streptococcus pneumoniae: comparison with PFGE and MLST.PLoS One. 2011;6(5):e19668. doi: 10.1371/journal.pone.0019668. Epub 2011 May 26. PLoS One. 2011. PMID: 21637335 Free PMC article.
-
[Molecular typing characterization of food-borne methicillin-resistant Staphylococcus aureus in China].Zhonghua Yu Fang Yi Xue Za Zhi. 2018 Apr 6;52(4):364-371. doi: 10.3760/cma.j.issn.0253-9624.2018.04.007. Zhonghua Yu Fang Yi Xue Za Zhi. 2018. PMID: 29614602 Chinese.
-
Multiple-locus variable-number tandem-repeat analysis of Streptococcus pneumoniae and comparison with multiple loci sequence typing.BMC Microbiol. 2012 Oct 22;12:241. doi: 10.1186/1471-2180-12-241. BMC Microbiol. 2012. PMID: 23088225 Free PMC article.
-
Comparison of fingerprinting methods for typing methicillin-resistant Staphylococcus aureus sequence type 398.J Clin Microbiol. 2009 Oct;47(10):3313-22. doi: 10.1128/JCM.00910-09. Epub 2009 Aug 26. J Clin Microbiol. 2009. PMID: 19710273 Free PMC article.
-
Comparison of classical multi-locus sequence typing software for next-generation sequencing data.Microb Genom. 2017 Jul 4;3(8):e000124. doi: 10.1099/mgen.0.000124. eCollection 2017 Aug. Microb Genom. 2017. PMID: 29026660 Free PMC article. Review.
Cited by
-
Russian isolates enlarge the known geographic diversity of Francisella tularensis subsp. mediasiatica.PLoS One. 2017 Sep 5;12(9):e0183714. doi: 10.1371/journal.pone.0183714. eCollection 2017. PLoS One. 2017. PMID: 28873421 Free PMC article.
-
Genotyping of French Bacillus anthracis strains based on 31-loci multi locus VNTR analysis: epidemiology, marker evaluation, and update of the internet genotype database.PLoS One. 2014 Jun 5;9(6):e95131. doi: 10.1371/journal.pone.0095131. eCollection 2014. PLoS One. 2014. PMID: 24901417 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

