Ddx46 is required for multi-lineage differentiation of hematopoietic stem cells in zebrafish
- PMID: 23635340
- PMCID: PMC3760052
- DOI: 10.1089/scd.2012.0623
Ddx46 is required for multi-lineage differentiation of hematopoietic stem cells in zebrafish
Abstract
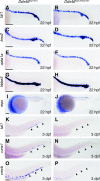
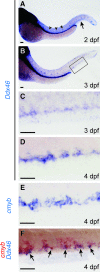
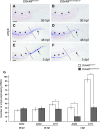
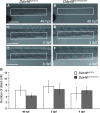
Balanced and precisely controlled processes between self-renewal and differentiation of hematopoietic stem cells (HSCs) into all blood lineages are critical for vertebrate definitive hematopoiesis. However, the molecular mechanisms underlying the maintenance and differentiation of HSCs have not been fully elucidated. Here, we show that zebrafish Ddx46, encoding a DEAD-box RNA helicase, is expressed in HSCs of the caudal hematopoietic tissue (CHT). The number of HSCs expressing the molecular markers cmyb or T-cell acute lymphocytic leukemia 1 (tal1) was markedly reduced in Ddx46 mutants. However, massive cell death of HSCs was not detected, and proliferation of HSCs was normal in the CHT of the mutants at 48 h postfertilization. We found that myelopoiesis occurred, but erythropoiesis and lymphopoiesis were suppressed, in Ddx46 mutants. Consistent with these results, the expression of spi1, encoding a regulator of myeloid development, was maintained, but the expression of gata1a, encoding a regulator of erythrocyte development, was downregulated in the mutants. Taken together, our results provide the first genetic evidence that zebrafish Ddx46 is required for the multilineage differentiation of HSCs during development, through the regulation of specific gene expressions.
Figures

Similar articles
-
DEAD-box protein Ddx46 is required for the development of the digestive organs and brain in zebrafish.PLoS One. 2012;7(3):e33675. doi: 10.1371/journal.pone.0033675. Epub 2012 Mar 19. PLoS One. 2012. PMID: 22442707 Free PMC article.
-
Distinct regulation of the anterior and posterior myeloperoxidase expression by Etv2 and Gata1 during primitive Granulopoiesis in zebrafish.Dev Biol. 2014 Sep 1;393(1):149-159. doi: 10.1016/j.ydbio.2014.06.011. Epub 2014 Jun 20. Dev Biol. 2014. PMID: 24956419 Free PMC article.
-
The zebrafish ETS transcription factor Fli1b functions upstream of Scl/Tal1 during embryonic hematopoiesis.Biol Open. 2025 Apr 15;14(4):bio061948. doi: 10.1242/bio.061948. Epub 2025 Apr 9. Biol Open. 2025. PMID: 40079219 Free PMC article.
-
SCL/TAL1 in Hematopoiesis and Cellular Reprogramming.Curr Top Dev Biol. 2016;118:163-204. doi: 10.1016/bs.ctdb.2016.01.004. Epub 2016 Feb 18. Curr Top Dev Biol. 2016. PMID: 27137657 Review.
-
Transcription Factor TAL1 in Erythropoiesis.Adv Exp Med Biol. 2024;1459:243-258. doi: 10.1007/978-3-031-62731-6_11. Adv Exp Med Biol. 2024. PMID: 39017847 Review.
Cited by
-
Knockdown of DDX46 Inhibits the Invasion and Tumorigenesis in Osteosarcoma Cells.Oncol Res. 2017 Mar 13;25(3):417-425. doi: 10.3727/096504016X14747253292210. Epub 2016 Sep 30. Oncol Res. 2017. PMID: 27697093 Free PMC article.
-
DDX46 silencing inhibits cell proliferation by activating apoptosis and autophagy in cutaneous squamous cell carcinoma.Mol Med Rep. 2020 Nov;22(5):4236-4242. doi: 10.3892/mmr.2020.11509. Epub 2020 Sep 14. Mol Med Rep. 2020. PMID: 33000271 Free PMC article.
-
Thyroid hormone deficiency during zebrafish development impairs central nervous system myelination.PLoS One. 2021 Aug 17;16(8):e0256207. doi: 10.1371/journal.pone.0256207. eCollection 2021. PLoS One. 2021. PMID: 34403440 Free PMC article.
-
The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus.Nat Immunol. 2017 Oct;18(10):1094-1103. doi: 10.1038/ni.3830. Epub 2017 Aug 28. Nat Immunol. 2017. PMID: 28846086
-
m6A eraser ALKBH5/treRNA1/DDX46 axis regulates BCR expression.Neoplasia. 2025 Apr;62:101144. doi: 10.1016/j.neo.2025.101144. Epub 2025 Feb 22. Neoplasia. 2025. PMID: 39987653 Free PMC article.
References
-
- Cumano A. Godin I. Ontogeny of the hematopoietic system. Annu Rev Immunol. 2007;25:745–785. - PubMed
-
- Medvinsky A. Rybtsov S. Taoudi S. Embryonic origin of the adult hematopoietic system: advances and questions. Development. 2011;138:1017–1031. - PubMed
-
- Paik EJ. Zon LI. Hematopoietic development in the zebrafish. Int J Dev Biol. 2010;54:1127–1137. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

