Evolutionary cell biology of chromosome segregation: insights from trypanosomes
- PMID: 23635522
- PMCID: PMC3866873
- DOI: 10.1098/rsob.130023
Evolutionary cell biology of chromosome segregation: insights from trypanosomes
Abstract
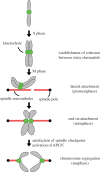
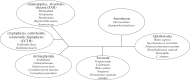
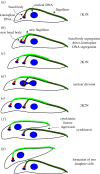
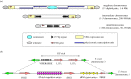
Faithful transmission of genetic material is essential for the survival of all organisms. Eukaryotic chromosome segregation is driven by the kinetochore that assembles onto centromeric DNA to capture spindle microtubules and govern the movement of chromosomes. Its molecular mechanism has been actively studied in conventional model eukaryotes, such as yeasts, worms, flies and human. However, these organisms are closely related in the evolutionary time scale and it therefore remains unclear whether all eukaryotes use a similar mechanism. The evolutionary origins of the segregation apparatus also remain enigmatic. To gain insights into these questions, it is critical to perform comparative studies. Here, we review our current understanding of the mitotic mechanism in Trypanosoma brucei, an experimentally tractable kinetoplastid parasite that branched early in eukaryotic history. No canonical kinetochore component has been identified, and the design principle of kinetochores might be fundamentally different in kinetoplastids. Furthermore, these organisms do not appear to possess a functional spindle checkpoint that monitors kinetochore-microtubule attachments. With these unique features and the long evolutionary distance from other eukaryotes, understanding the mechanism of chromosome segregation in T. brucei should reveal fundamental requirements for the eukaryotic segregation machinery, and may also provide hints about the origin and evolution of the segregation apparatus.
Keywords: CENP-A; Trypanosoma brucei; chromosome segregation; kinetochores; kinetoplastids; mitosis.
Figures

References
-
- Robinson NP, Bell SD. 2005. Origins of DNA replication in the three domains of life. FEBS J. 272, 3757–3766 10.1111/j.1742-4658.2005.04768.x (doi:10.1111/j.1742-4658.2005.04768.x) - DOI - PubMed
-
- Mott ML, Berger JM. 2007. DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 5, 343–354 10.1038/nrmicro1640 (doi:10.1038/nrmicro1640) - DOI - PubMed
-
- Gerdes K, Howard M, Szardenings F. 2010. Pushing and pulling in prokaryotic DNA segregation. Cell 141, 927–942 10.1016/j.cell.2010.05.033 (doi:10.1016/j.cell.2010.05.033) - DOI - PubMed
-
- Toro E, Shapiro L. 2010. Bacterial chromosome organization and segregation. Cold Spring Harb. Perspect. Biol. 2, a000349. 10.1101/cshperspect.a000349 (doi:10.1101/cshperspect.a000349) - DOI - PMC - PubMed
-
- Kalliomaa-Sanford AK, Rodriguez-Castañeda FA, McLeod BN, Latorre-Roselló V, Smith JH, Reimann J, Albers SV, Barillà D. 2012. Chromosome segregation in Archaea mediated by a hybrid DNA partition machine. Proc. Natl Acad. Sci. USA 109, 3754–3759 10.1073/pnas.1113384109 (doi:10.1073/pnas.1113384109) - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
