Approaches to querying bacterial genomes with transposon-insertion sequencing
- PMID: 23635712
- PMCID: PMC3849164
- DOI: 10.4161/rna.24765
Approaches to querying bacterial genomes with transposon-insertion sequencing
Abstract
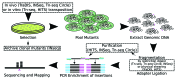
In this review, we discuss transposon-insertion sequencing, variously known in the literature as TraDIS, Tn-seq, INSeq, and HITS. By monitoring a large library of single transposon-insertion mutants with high-throughput sequencing, these methods can rapidly identify genomic regions that contribute to organismal fitness under any condition assayable in the laboratory with exquisite resolution. We discuss the various protocols that have been developed and methods for analysis. We provide an overview of studies that have examined the reproducibility and accuracy of these methods, as well as studies showing the advantages offered by the high resolution and dynamic range of high-throughput sequencing over previous methods. We review a number of applications in the literature, from predicting genes essential for in vitro growth to directly assaying requirements for survival under infective conditions in vivo. We also highlight recent progress in assaying non-coding regions of the genome in addition to known coding sequences, including the combining of RNA-seq with high-throughput transposon mutagenesis.
Keywords: bacteria; essential genes; sRNA; sequencing; systems biology; transposon mutagenesis.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
