Lung tumor NF-κB signaling promotes T cell-mediated immune surveillance
- PMID: 23635779
- PMCID: PMC3668836
- DOI: 10.1172/JCI67250
Lung tumor NF-κB signaling promotes T cell-mediated immune surveillance
Abstract
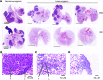
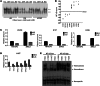
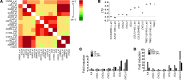
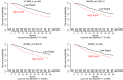
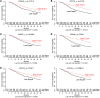
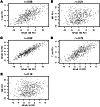
NF-κB is constitutively activated in many cancer types and is a potential key mediator of tumor-associated inflammation, tumor growth, and metastasis. We investigated the role of cancer cell NF-κB activity in T cell-mediated antitumor responses. In tumors rendered immunogenic by model antigen expression or following administration of antitumor vaccines, we found that high NF-κB activity leads to tumor rejection and/or growth suppression in mice. Using a global RNA expression microarray, we demonstrated that NF-κB enhanced expression of several T cell chemokines, including Ccl2, and decreased CCL2 expression was associated with enhanced tumor growth in a mouse lung cancer model. To investigate NF-κB function in human lung tumors, we identified a gene expression signature in human lung adenocarcinoma cell lines that was associated with NF-κB activity level. In patient tumor samples, overall lung tumor NF-κB activity was strongly associated with T cell infiltration but not with cancer cell proliferation. These results therefore indicate that NF-κB activity mediates immune surveillance and promotes antitumor T cell responses in both murine and human lung cancer.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

