PT-ACRAMTU, a platinum-acridine anticancer agent, lengthens and aggregates, but does not stiffen or soften DNA
- PMID: 23636685
- PMCID: PMC3767762
- DOI: 10.1007/s12013-013-9614-8
PT-ACRAMTU, a platinum-acridine anticancer agent, lengthens and aggregates, but does not stiffen or soften DNA
Abstract
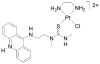
We used atomic force microscopy (AFM) to study the dose-dependent change in conformational and mechanical properties of DNA treated with PT-ACRAMTU ([PtCl(en)(ACRAMTU-S)](NO3)2, (en = ethane-1,2-diamine, ACRAMTU = 1-[2-(acridin-9-ylamino)ethyl]-1,3-dimethylthiourea. PT-ACRAMTU is the parent drug of a family of non-classical platinum-based agents that show potent activity in non-small cell lung cancer in vitro and in vivo. Its acridine moiety intercalates between DNA bases, while the platinum group forms mono-adducts with DNA bases. AFM images show that PT-ACRAMTU causes some DNA looping and aggregation at drug-to-base pair ratio (r b) of 0.1 and higher. Very significant lengthening of the DNA was observed with increasing doses of PT-ACRAMTU, and reached saturation at an r b of 0.15. At r b of 0.1, lengthening was 0.6 nm per drug molecule, which is more than one fully stretched base pair stack can accommodate, indicating that ACRAMTU also disturbs the stacking of neighboring base pair stacks. Analysis of the AFM images based on the worm-like chain (WLC) model showed that PT-ACRAMTU did not change the flexibility of (non-aggregated) DNA, despite the extreme lengthening. The persistence length of untreated DNA and DNA treated with PT-ACRAMTU was in the range of 49-65 nm. Potential consequences of the perturbations caused by this agent for the recognition and processing of the DNA adducts it forms are discussed.
Figures





Similar articles
-
Solution structural study of a DNA duplex containing the guanine-N7 adduct formed by a cytotoxic platinum-acridine hybrid agent.Biochemistry. 2005 Apr 26;44(16):6059-70. doi: 10.1021/bi050021b. Biochemistry. 2005. PMID: 15835895
-
Unprecedented monofunctional metalation of adenine nucleobase in guanine- and thymine-containing dinucleotide sequences by a cytotoxic platinum-acridine hybrid agent.J Am Chem Soc. 2003 Aug 13;125(32):9629-37. doi: 10.1021/ja0351443. J Am Chem Soc. 2003. PMID: 12904029
-
DNA minor groove adducts formed by a platinum-acridine conjugate inhibit association of tata-binding protein with its cognate sequence.Biochemistry. 2005 Aug 23;44(33):11262-8. doi: 10.1021/bi050745n. Biochemistry. 2005. PMID: 16101310
-
Adenine-N3 in the DNA minor groove - an emerging target for platinum containing anticancer pharmacophores.Anticancer Agents Med Chem. 2007 Jan;7(1):125-38. doi: 10.2174/187152007779313991. Anticancer Agents Med Chem. 2007. PMID: 17266509 Review.
-
[Drug discovery research in in-vivo antitumor-active azolato-bridged dinuclear Pt(II) complexes].Yakugaku Zasshi. 2012;132(3):253-9. doi: 10.1248/yakushi.132.253. Yakugaku Zasshi. 2012. PMID: 22382827 Review. Japanese.
Cited by
-
AFM of self-assembled lambda DNA-histone networks.Colloids Surf B Biointerfaces. 2015 Oct 1;134:17-25. doi: 10.1016/j.colsurfb.2015.06.026. Epub 2015 Jun 19. Colloids Surf B Biointerfaces. 2015. PMID: 26141439 Free PMC article.
-
Design and cellular studies of a carbon nanotube-based delivery system for a hybrid platinum-acridine anticancer agent.J Inorg Biochem. 2016 Dec;165:170-180. doi: 10.1016/j.jinorgbio.2016.07.016. Epub 2016 Jul 27. J Inorg Biochem. 2016. PMID: 27496614 Free PMC article.
-
Dual Photoreactivity of a New Rh2(II,II) Complex for Biological Applications.Inorganica Chim Acta. 2017 Jan 1;454:149-154. doi: 10.1016/j.ica.2016.04.001. Epub 2016 Apr 18. Inorganica Chim Acta. 2017. PMID: 30026633 Free PMC article.
References
-
- Eastman A. Activation of programmed cell death by anticancer agents - cisplatin as a model system. Cancer Cells-a Monthly Review. 1990;2:275–280. - PubMed
-
- Chaney SG, Campbell SL, Temple B, Bassett E, Wu YB, Faldu M. Protein interactions with platinum-DNA adducts: from structure to function. Journal of Inorganic Biochemistry. 2004;98:1551–1559. - PubMed
-
- Kartalou M, Essigmann JM. Mechanisms of resistance to cisplatin. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis. 2001;478:23–43. - PubMed
-
- Takahara PM, Frederick CA, Lippard SJ. Crystal structure of the anticancer drug cisplatin bound to duplex DNA. Journal of the American Chemical Society. 1996;118:12309–12321.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

