Enhancer transcripts mark active estrogen receptor binding sites
- PMID: 23636943
- PMCID: PMC3730096
- DOI: 10.1101/gr.152306.112
Enhancer transcripts mark active estrogen receptor binding sites
Abstract
We have integrated and analyzed a large number of data sets from a variety of genomic assays using a novel computational pipeline to provide a global view of estrogen receptor 1 (ESR1; a.k.a. ERα) enhancers in MCF-7 human breast cancer cells. Using this approach, we have defined a class of primary transcripts (eRNAs) that are transcribed uni- or bidirectionally from estrogen receptor binding sites (ERBSs) with an average transcription unit length of ∼3-5 kb. The majority are up-regulated by short treatments with estradiol (i.e., 10, 25, or 40 min) with kinetics that precede or match the induction of the target genes. The production of eRNAs at ERBSs is strongly correlated with the enrichment of a number of genomic features that are associated with enhancers (e.g., H3K4me1, H3K27ac, EP300/CREBBP, RNA polymerase II, open chromatin architecture), as well as enhancer looping to target gene promoters. In the absence of eRNA production, strong enrichment of these features is not observed, even though ESR1 binding is evident. We find that flavopiridol, a CDK9 inhibitor that blocks transcription elongation, inhibits eRNA production but does not affect other molecular indicators of enhancer activity, suggesting that eRNA production occurs after the assembly of active enhancers. Finally, we show that an enhancer transcription "signature" based on GRO-seq data can be used for de novo enhancer prediction across cell types. Together, our studies shed new light on the activity of ESR1 at its enhancer sites and provide new insights about enhancer function.
Figures
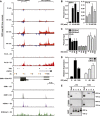




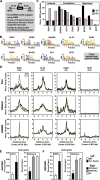
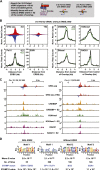
References
-
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
