Mapping functional transcription factor networks from gene expression data
- PMID: 23636944
- PMCID: PMC3730105
- DOI: 10.1101/gr.150904.112
Mapping functional transcription factor networks from gene expression data
Abstract
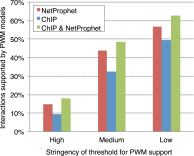
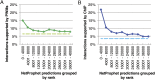
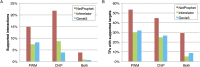
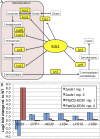
A critical step in understanding how a genome functions is determining which transcription factors (TFs) regulate each gene. Accordingly, extensive effort has been devoted to mapping TF networks. In Saccharomyces cerevisiae, protein-DNA interactions have been identified for most TFs by ChIP-chip, and expression profiling has been done on strains deleted for most TFs. These studies revealed that there is little overlap between the genes whose promoters are bound by a TF and those whose expression changes when the TF is deleted, leaving us without a definitive TF network for any eukaryote and without an efficient method for mapping functional TF networks. This paper describes NetProphet, a novel algorithm that improves the efficiency of network mapping from gene expression data. NetProphet exploits a fundamental observation about the nature of TF networks: The response to disrupting or overexpressing a TF is strongest on its direct targets and dissipates rapidly as it propagates through the network. Using S. cerevisiae data, we show that NetProphet can predict thousands of direct, functional regulatory interactions, using only gene expression data. The targets that NetProphet predicts for a TF are at least as likely to have sites matching the TF's binding specificity as the targets implicated by ChIP. Unlike most ChIP targets, the NetProphet targets also show evidence of functional regulation. This suggests a surprising conclusion: The best way to begin mapping direct, functional TF-promoter interactions may not be by measuring binding. We also show that NetProphet yields new insights into the functions of several yeast TFs, including a well-studied TF, Cbf1, and a completely unstudied TF, Eds1.
Figures

References
-
- Abdulrehman D, Monteiro PT, Teixeira MC, Mira NP, Lourenco AB, dos Santos SC, Cabrito TR, Francisco AP, Madeira SC, Aires RS, et al. 2011. YEASTRACT: Providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface. Nucleic Acids Res 39: D136–D140 - PMC - PubMed
-
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA 2004. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol 14: 283–291 - PubMed
-
- Balaji S, Babu MM, Iyer LM, Luscombe NM, Aravind L 2006. Comprehensive analysis of combinatorial regulation using the transcriptional regulatory network of yeast. J Mol Biol 360: 213–227 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
