Repair of double-strand breaks by end joining
- PMID: 23637284
- PMCID: PMC3632057
- DOI: 10.1101/cshperspect.a012757
Repair of double-strand breaks by end joining
Abstract
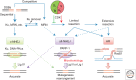
Nonhomologous end joining (NHEJ) refers to a set of genome maintenance pathways in which two DNA double-strand break (DSB) ends are (re)joined by apposition, processing, and ligation without the use of extended homology to guide repair. Canonical NHEJ (c-NHEJ) is a well-defined pathway with clear roles in protecting the integrity of chromosomes when DSBs arise. Recent advances have revealed much about the identity, structure, and function of c-NHEJ proteins, but many questions exist regarding their concerted action in the context of chromatin. Alternative NHEJ (alt-NHEJ) refers to more recently described mechanism(s) that repair DSBs in less-efficient backup reactions. There is great interest in defining alt-NHEJ more precisely, including its regulation relative to c-NHEJ, in light of evidence that alt-NHEJ can execute chromosome rearrangements. Progress toward these goals is reviewed.
Figures



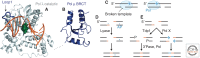
References
-
- Ahnesorg P, Smith P, Jackson SP 2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124: 301–313 - PubMed
-
- Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS 2007. Crystal structure of human XLF: A twist in nonhomologous DNA end-joining. Mol Cell 28: 1093–1101 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
