Rescuing stalled or damaged replication forks
- PMID: 23637285
- PMCID: PMC3632063
- DOI: 10.1101/cshperspect.a012815
Rescuing stalled or damaged replication forks
Abstract
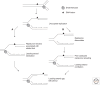
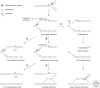
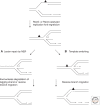
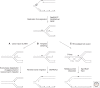
In recent years, an increasing number of studies have shown that prokaryotes and eukaryotes are armed with sophisticated mechanisms to restart stalled or collapsed replication forks. Although these processes are better understood in bacteria, major breakthroughs have also been made to explain how fork restart mechanisms operate in eukaryotic cells. In particular, repriming on the leading strand and fork regression are now established as critical for the maintenance and recovery of stalled forks in both systems. Despite the lack of conservation between the factors involved, these mechanisms are strikingly similar in eukaryotes and prokaryotes. However, they differ in that fork restart occurs in the context of chromatin in eukaryotes and is controlled by multiple regulatory pathways.
Figures
References
-
- Aguilera A, Gomez-Gonzalez B 2008. Genome instability: A mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
