Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry
- PMID: 23637301
- PMCID: PMC3697718
- DOI: 10.1128/JCM.00492-13
Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry
Abstract
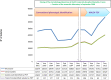
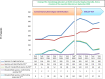
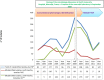
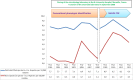
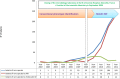
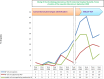
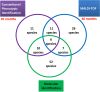
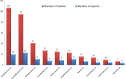
During the past 5 years, matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) has become a powerful tool for routine identification in many clinical laboratories. We analyzed our 11-year experience in routine identification of clinical isolates (40 months using MALDI-TOF MS and 91 months using conventional phenotypic identification [CPI]). Among the 286,842 clonal isolates, 284,899 isolates of 459 species were identified. The remaining 1,951 isolates were misidentified and required confirmation using a second phenotypic identification for 670 isolates and using a molecular technique for 1,273 isolates of 339 species. MALDI-TOF MS annually identified 112 species, i.e., 36 species/10,000 isolates, compared to 44 species, i.e., 19 species/10,000 isolates, for CPI. Only 50 isolates required second phenotypic identifications during the MALDI-TOF MS period (i.e., 4.5 reidentifications/10,000 isolates) compared with 620 isolates during the CPI period (i.e., 35.2/10,000 isolates). We identified 128 bacterial species rarely reported as human pathogens, including 48 using phenotypic techniques (22 using CPI and 37 using MALDI-TOF MS). Another 75 rare species were identified using molecular methods. MALDI-TOF MS reduced the time required for identification by 55-fold and 169-fold and the cost by 5-fold and 96-fold compared with CPI and gene sequencing, respectively. MALDI-TOF MS was a powerful tool not only for routine bacterial identification but also for identification of rare bacterial species implicated in human infectious diseases. The ability to rapidly identify bacterial species rarely described as pathogens in specific clinical specimens will help us to study the clinical burden resulting from the emergence of these species as human pathogens, and MALDI-TOF MS may be considered an alternative to molecular methods in clinical laboratories.
Figures
References
-
- Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. 2008. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 14:908–934 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

