Cellular chaperonin CCTγ contributes to rabies virus replication during infection
- PMID: 23637400
- PMCID: PMC3700271
- DOI: 10.1128/JVI.03186-12
Cellular chaperonin CCTγ contributes to rabies virus replication during infection
Abstract
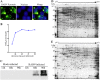
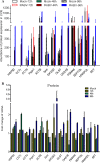
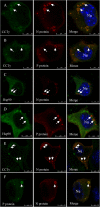
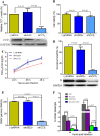
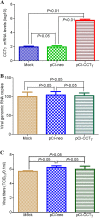
Rabies, as the oldest known infectious disease, remains a serious threat to public health worldwide. The eukaryotic cytosolic chaperonin TRiC/CCT complex facilitates the folding of proteins through ATP hydrolysis. Here, we investigated the expression, cellular localization, and function of neuronal CCTγ during neurotropic rabies virus (RABV) infection using mouse N2a cells as a model. Following RABV infection, 24 altered proteins were identified by using two-dimensional electrophoresis and mass spectrometry, including 20 upregulated proteins and 4 downregulated proteins. In mouse N2a cells infected with RABV or cotransfected with RABV genes encoding nucleoprotein (N) and phosphoprotein (P), confocal microscopy demonstrated that upregulated cellular CCTγ was colocalized with viral proteins N and P, which formed a hollow cricoid inclusion within the region around the nucleus. These inclusions, which correspond to Negri bodies (NBs), did not form in mouse N2a cells only expressing the viral protein N or P. Knockdown of CCTγ by lentivirus-mediated RNA interference led to significant inhibition of RABV replication. These results demonstrate that the complex consisting of viral proteins N and P recruits CCTγ to NBs and identify the chaperonin CCTγ as a host factor that facilitates intracellular RABV replication. This work illustrates how viruses can utilize cellular chaperonins and compartmentalization for their own benefit.
Figures

References
-
- Lafon M. 2011. Evasive strategies in rabies virus infection. Adv. Virus Res. 79:33–53 - PubMed
-
- Jamin M, Leyrat C, Ribeiro EA, Gerard FCA, Ivanov I, Ruigrok RWH. 2011. Structure, interactions with host cell and functions of rhabdovirus phosphoprotein. Future Virol. 6:465–481
-
- WHO September 2012. Rabies: fact sheet no. 99. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs099/en/ Accessed 26 December 2012
-
- Negri A. 1903. Contributo allo studio dell'eziologia della rabbia. Boll. Soc. Med. Chir. Pavia 2:88–115
-
- Suchy A, Bauder B, Gelbmann W, Lohr CV, Teifke JP, Weissenbock H. 2000. Diagnosis of feline herpesvirus infection by immunohistochemistry, polymerase chain reaction, and in situ hybridization. J. Vet. Diagn. Invest. 12:186–191 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

