Six homeoproteins directly activate Myod expression in the gene regulatory networks that control early myogenesis
- PMID: 23637613
- PMCID: PMC3636133
- DOI: 10.1371/journal.pgen.1003425
Six homeoproteins directly activate Myod expression in the gene regulatory networks that control early myogenesis
Abstract
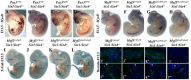
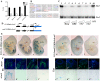
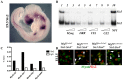
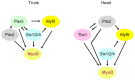
In mammals, several genetic pathways have been characterized that govern engagement of multipotent embryonic progenitors into the myogenic program through the control of the key myogenic regulatory gene Myod. Here we demonstrate the involvement of Six homeoproteins. We first targeted into a Pax3 allele a sequence encoding a negative form of Six4 that binds DNA but cannot interact with essential Eya co-factors. The resulting embryos present hypoplasic skeletal muscles and impaired Myod activation in the trunk in the absence of Myf5/Mrf4. At the axial level, we further show that Myod is still expressed in compound Six1/Six4:Pax3 but not in Six1/Six4:Myf5 triple mutant embryos, demonstrating that Six1/4 participates in the Pax3-Myod genetic pathway. Myod expression and head myogenesis is preserved in Six1/Six4:Myf5 triple mutant embryos, illustrating that upstream regulators of Myod in different embryonic territories are distinct. We show that Myod regulatory regions are directly controlled by Six proteins and that, in the absence of Six1 and Six4, Six2 can compensate.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo.Development. 2005 May;132(9):2235-49. doi: 10.1242/dev.01773. Epub 2005 Mar 23. Development. 2005. PMID: 15788460
-
Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo.Dev Biol. 2007 Feb 15;302(2):602-16. doi: 10.1016/j.ydbio.2006.08.059. Epub 2006 Sep 1. Dev Biol. 2007. PMID: 17098221
-
Six and Eya expression during human somitogenesis and MyoD gene family activation.J Muscle Res Cell Motil. 2002;23(3):255-64. doi: 10.1023/a:1020990825644. J Muscle Res Cell Motil. 2002. PMID: 12500905
-
How is myogenesis initiated in chordates?Folia Biol (Krakow). 2012;60(3-4):107-19. doi: 10.3409/fb60_3-4.107-119. Folia Biol (Krakow). 2012. PMID: 23342904 Review.
-
Gene regulatory networks and transcriptional mechanisms that control myogenesis.Dev Cell. 2014 Feb 10;28(3):225-38. doi: 10.1016/j.devcel.2013.12.020. Dev Cell. 2014. PMID: 24525185 Review.
Cited by
-
MyoD reprogramming requires Six1 and Six4 homeoproteins: genome-wide cis-regulatory module analysis.Nucleic Acids Res. 2016 Oct 14;44(18):8621-8640. doi: 10.1093/nar/gkw512. Epub 2016 Jun 14. Nucleic Acids Res. 2016. PMID: 27302134 Free PMC article.
-
The emergence of Pax7-expressing muscle stem cells during vertebrate head muscle development.Front Aging Neurosci. 2015 May 19;7:62. doi: 10.3389/fnagi.2015.00062. eCollection 2015. Front Aging Neurosci. 2015. PMID: 26042028 Free PMC article.
-
Six1 homeoprotein drives myofiber type IIA specialization in soleus muscle.Skelet Muscle. 2016 Sep 5;6(1):30. doi: 10.1186/s13395-016-0102-x. eCollection 2016. Skelet Muscle. 2016. PMID: 27597886 Free PMC article.
-
Master regulators of skeletal muscle lineage development and pluripotent stem cells differentiation.Cell Regen. 2021 Oct 1;10(1):31. doi: 10.1186/s13619-021-00093-5. Cell Regen. 2021. PMID: 34595600 Free PMC article. Review.
-
Dynamics of transcriptional (re)-programming of syncytial nuclei in developing muscles.BMC Biol. 2017 Jun 9;15(1):48. doi: 10.1186/s12915-017-0386-2. BMC Biol. 2017. PMID: 28599653 Free PMC article.
References
-
- Gehring WJ, Ikeo K (1999) Pax 6: mastering eye morphogenesis and eye evolution. Trends in genetics 15: 371–377. - PubMed
-
- van Heyningen V, Williamson KA (2002) PAX6 in sensory development. Human molecular genetics 11: 1161–1167. - PubMed
-
- Jemc J, Rebay I (2007) The eyes absent family of phosphotyrosine phosphatases: properties and roles in developmental regulation of transcription. Annual review of biochemistry 76: 513–538. - PubMed
-
- Buckingham M, Relaix F (2007) The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annual review of cell and developmental biology 23: 645–673. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

