Disruption of TTDA results in complete nucleotide excision repair deficiency and embryonic lethality
- PMID: 23637614
- PMCID: PMC3630102
- DOI: 10.1371/journal.pgen.1003431
Disruption of TTDA results in complete nucleotide excision repair deficiency and embryonic lethality
Abstract
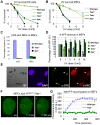
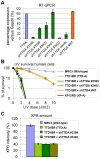
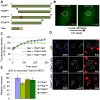
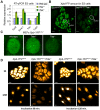
The ten-subunit transcription factor IIH (TFIIH) plays a crucial role in transcription and nucleotide excision repair (NER). Inactivating mutations in the smallest 8-kDa TFB5/TTDA subunit cause the neurodevelopmental progeroid repair syndrome trichothiodystrophy A (TTD-A). Previous studies have shown that TTDA is the only TFIIH subunit that appears not to be essential for NER, transcription, or viability. We studied the consequences of TTDA inactivation by generating a Ttda knock-out (Ttda(-/-) ) mouse-model resembling TTD-A patients. Unexpectedly, Ttda(-/-) mice were embryonic lethal. However, in contrast to full disruption of all other TFIIH subunits, viability of Ttda(-/-) cells was not affected. Surprisingly, Ttda(-/-) cells were completely NER deficient, contrary to the incomplete NER deficiency of TTD-A patient-derived cells. We further showed that TTD-A patient mutations only partially inactivate TTDA function, explaining the relatively mild repair phenotype of TTD-A cells. Moreover, Ttda(-/-) cells were also highly sensitive to oxidizing agents. These findings reveal an essential role of TTDA for life, nucleotide excision repair, and oxidative DNA damage repair and identify Ttda(-/-) cells as a unique class of TFIIH mutants.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374. - PubMed
-
- Sugasawa K, Akagi J, Nishi R, Iwai S, Hanaoka F (2009) Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning. Mol Cell 36: 642–653. - PubMed
-
- Conaway RC, Conaway JW (1993) General initiation factors for RNA polymerase II. Annu Rev Biochem 62: 161–190. - PubMed
-
- Egly JM (2001) The 14th Datta Lecture. TFIIH: from transcription to clinic. FEBS Lett 498: 124–128. - PubMed
-
- Zurita M, Merino C (2003) The transcriptional complexity of the TFIIH complex. Trends Genet 19: 578–584. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

