Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases
- PMID: 23637617
- PMCID: PMC3630097
- DOI: 10.1371/journal.pgen.1003439
Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases
Abstract
DNA methylation is an epigenetic modification that plays a crucial role in normal mammalian development, retrotransposon silencing, and cellular reprogramming. Although methylation mainly occurs on the cytosine in a CG site, non-CG methylation is prevalent in pluripotent stem cells, brain, and oocytes. We previously identified non-CG methylation in several CG-rich regions in mouse germinal vesicle oocytes (GVOs), but the overall distribution of non-CG methylation and the enzymes responsible for this modification are unknown. Using amplification-free whole-genome bisulfite sequencing, which can be used with minute amounts of DNA, we constructed the base-resolution methylome maps of GVOs, non-growing oocytes (NGOs), and mutant GVOs lacking the DNA methyltransferase Dnmt1, Dnmt3a, Dnmt3b, or Dnmt3L. We found that nearly two-thirds of all methylcytosines occur in a non-CG context in GVOs. The distribution of non-CG methylation closely resembled that of CG methylation throughout the genome and showed clear enrichment in gene bodies. Compared to NGOs, GVOs were over four times more methylated at non-CG sites, indicating that non-CG methylation accumulates during oocyte growth. Lack of Dnmt3a or Dnmt3L resulted in a global reduction in both CG and non-CG methylation, showing that non-CG methylation depends on the Dnmt3a-Dnmt3L complex. Dnmt3b was dispensable. Of note, lack of Dnmt1 resulted in a slight decrease in CG methylation, suggesting that this maintenance enzyme plays a role in non-dividing oocytes. Dnmt1 may act on CG sites that remain hemimethylated in the de novo methylation process. Our results provide a basis for understanding the mechanisms and significance of non-CG methylation in mammalian oocytes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
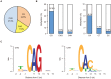
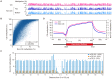
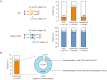
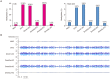
Similar articles
-
Role of UHRF1 in de novo DNA methylation in oocytes and maintenance methylation in preimplantation embryos.PLoS Genet. 2017 Oct 4;13(10):e1007042. doi: 10.1371/journal.pgen.1007042. eCollection 2017 Oct. PLoS Genet. 2017. PMID: 28976982 Free PMC article.
-
Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks.PLoS Genet. 2012 Jan;8(1):e1002440. doi: 10.1371/journal.pgen.1002440. Epub 2012 Jan 5. PLoS Genet. 2012. PMID: 22242016 Free PMC article.
-
DNMT1, DNMT3A and DNMT3B proteins are differently expressed in mouse oocytes and early embryos.J Mol Histol. 2017 Dec;48(5-6):417-426. doi: 10.1007/s10735-017-9739-y. Epub 2017 Oct 13. J Mol Histol. 2017. PMID: 29027601
-
DNA Methyltransferases in Mammalian Oocytes.Results Probl Cell Differ. 2017;63:211-222. doi: 10.1007/978-3-319-60855-6_10. Results Probl Cell Differ. 2017. PMID: 28779320 Review.
-
Dynamic expression of DNA methyltransferases (DNMTs) in oocytes and early embryos.Biochimie. 2015 Sep;116:103-13. doi: 10.1016/j.biochi.2015.06.019. Epub 2015 Jul 2. Biochimie. 2015. PMID: 26143007 Review.
Cited by
-
NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing.Nat Genet. 2020 Oct;52(10):1088-1098. doi: 10.1038/s41588-020-0689-z. Epub 2020 Sep 14. Nat Genet. 2020. PMID: 32929285
-
Impact of neonatal iron deficiency on hippocampal DNA methylation and gene transcription in a porcine biomedical model of cognitive development.BMC Genomics. 2016 Nov 3;17(1):856. doi: 10.1186/s12864-016-3216-y. BMC Genomics. 2016. PMID: 27809765 Free PMC article.
-
Tet enzymes are essential for early embryogenesis and completion of embryonic genome activation.EMBO Rep. 2022 Feb 3;23(2):e53968. doi: 10.15252/embr.202153968. Epub 2021 Dec 6. EMBO Rep. 2022. PMID: 34866320 Free PMC article.
-
DNA Methylation Reprogramming during Mammalian Development.Genes (Basel). 2019 Mar 29;10(4):257. doi: 10.3390/genes10040257. Genes (Basel). 2019. PMID: 30934924 Free PMC article. Review.
-
Overexpression of Human-Derived DNMT3A Induced Intergenerational Inheritance of Active DNA Methylation Changes in Rat Sperm.Front Genet. 2017 Dec 12;8:207. doi: 10.3389/fgene.2017.00207. eCollection 2017. Front Genet. 2017. PMID: 29312436 Free PMC article.
References
-
- Sasaki H, Matsui Y (2008) Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet 9: 129–140. - PubMed
-
- Laird PW (2010) Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet 11: 191–203. - PubMed
-
- Krueger F, Kreck B, Franke A, Andrews SR (2012) DNA methylome analysis using short bisulfite sequencing data. Nat Methods 9: 145–151. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

