Signatures of diversifying selection in European pig breeds
- PMID: 23637623
- PMCID: PMC3636142
- DOI: 10.1371/journal.pgen.1003453
Signatures of diversifying selection in European pig breeds
Abstract
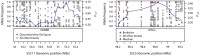
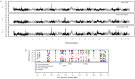
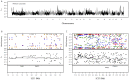
Following domestication, livestock breeds have experienced intense selection pressures for the development of desirable traits. This has resulted in a large diversity of breeds that display variation in many phenotypic traits, such as coat colour, muscle composition, early maturity, growth rate, body size, reproduction, and behaviour. To better understand the relationship between genomic composition and phenotypic diversity arising from breed development, the genomes of 13 traditional and commercial European pig breeds were scanned for signatures of diversifying selection using the Porcine60K SNP chip, applying a between-population (differentiation) approach. Signatures of diversifying selection between breeds were found in genomic regions associated with traits related to breed standard criteria, such as coat colour and ear morphology. Amino acid differences in the EDNRB gene appear to be associated with one of these signatures, and variation in the KITLG gene may be associated with another. Other selection signals were found in genomic regions including QTLs and genes associated with production traits such as reproduction, growth, and fat deposition. Some selection signatures were associated with regions showing evidence of introgression from Asian breeds. When the European breeds were compared with wild boar, genomic regions with high levels of differentiation harboured genes related to bone formation, growth, and fat deposition.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Larson G, Dobney K, Albarella U, Fang MY, Matisoo-Smith E, et al. (2005) Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 307: 1618–1621. - PubMed
-
- FAO (2007) The State of the World's Animal Genetic Resources for Food and Agriculture – in brief. Commission on Genetic Resources for Food and Agriculture: FAO, Rome, Italy.
-
- Darwin C (1868) The Variation of Animals and Plants under Domestication. London: John Murray.
-
- BPA (2002) British Pig Breeds. Trumpington, Cambridge: British Pig Association.
Publication types
MeSH terms
Grants and funding
- BBS/E/D/20211550/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/05191133/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211554/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_PC_U127527200/MRC_/Medical Research Council/United Kingdom
- MC_PC_U127592696/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

