SNF5 is an essential executor of epigenetic regulation during differentiation
- PMID: 23637628
- PMCID: PMC3636213
- DOI: 10.1371/journal.pgen.1003459
SNF5 is an essential executor of epigenetic regulation during differentiation
Abstract
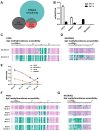
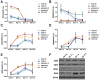
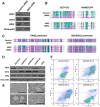
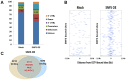
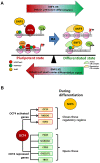
Nucleosome occupancy controls the accessibility of the transcription machinery to DNA regulatory regions and serves an instructive role for gene expression. Chromatin remodelers, such as the BAF complexes, are responsible for establishing nucleosome occupancy patterns, which are key to epigenetic regulation along with DNA methylation and histone modifications. Some reports have assessed the roles of the BAF complex subunits and stemness in murine embryonic stem cells. However, the details of the relationships between remodelers and transcription factors in altering chromatin configuration, which ultimately affects gene expression during cell differentiation, remain unclear. Here for the first time we demonstrate that SNF5, a core subunit of the BAF complex, negatively regulates OCT4 levels in pluripotent cells and is essential for cell survival during differentiation. SNF5 is responsible for generating nucleosome-depleted regions (NDRs) at the regulatory sites of OCT4 repressed target genes such as PAX6 and NEUROG1, which are crucial for cell fate determination. Concurrently, SNF5 closes the NDRs at the regulatory regions of OCT4-activated target genes such as OCT4 itself and NANOG. Furthermore, using loss- and gain-of-function experiments followed by extensive genome-wide analyses including gene expression microarrays and ChIP-sequencing, we highlight that SNF5 plays dual roles during differentiation by antagonizing the expression of genes that were either activated or repressed by OCT4, respectively. Together, we demonstrate that SNF5 executes the switch between pluripotency and differentiation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Reik W (2007) Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425–432. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

