The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3
- PMID: 23637629
- PMCID: PMC3630093
- DOI: 10.1371/journal.pgen.1003461
The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3
Abstract
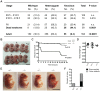
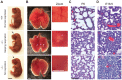
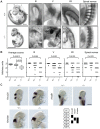
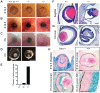
Embryonic development is tightly regulated by transcription factors and chromatin-associated proteins. H3K4me3 is associated with active transcription and H3K27me3 with gene repression, while the combination of both keeps genes required for development in a plastic state. Here we show that deletion of the H3K4me2/3 histone demethylase Jarid1b (Kdm5b/Plu1) results in major neonatal lethality due to respiratory failure. Jarid1b knockout embryos have several neural defects including disorganized cranial nerves, defects in eye development, and increased incidences of exencephaly. Moreover, in line with an overlap of Jarid1b and Polycomb target genes, Jarid1b knockout embryos display homeotic skeletal transformations typical for Polycomb mutants, supporting a functional interplay between Polycomb proteins and Jarid1b. To understand how Jarid1b regulates mouse development, we performed a genome-wide analysis of histone modifications, which demonstrated that normally inactive genes encoding developmental regulators acquire aberrant H3K4me3 during early embryogenesis in Jarid1b knockout embryos. H3K4me3 accumulates as embryonic development proceeds, leading to increased expression of neural master regulators like Pax6 and Otx2 in Jarid1b knockout brains. Taken together, these results suggest that Jarid1b regulates mouse development by protecting developmental genes from inappropriate acquisition of active histone modifications.
Conflict of interest statement
KH is a cofounder of EpiTherapeutics and has shares and warrants in the company. All other authors have declared that no competing financial interests exist.
Figures


References
-
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, et al. (2007) High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 129: 823–837. - PubMed
-
- Hublitz P, Albert M, Peters AHFM (2009) Mechanisms of transcriptional repression by histone lysine methylation. Int J Dev Biol 53: 335–354. - PubMed
-
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ (1995) Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378: 505–508. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

