Identification of a tissue-selective heat shock response regulatory network
- PMID: 23637632
- PMCID: PMC3630107
- DOI: 10.1371/journal.pgen.1003466
Identification of a tissue-selective heat shock response regulatory network
Abstract
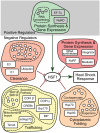
The heat shock response (HSR) is essential to survive acute proteotoxic stress and has been studied extensively in unicellular organisms and tissue culture cells, but to a lesser extent in intact metazoan animals. To identify the regulatory pathways that control the HSR in Caenorhabditis elegans, we performed a genome-wide RNAi screen and identified 59 genes corresponding to 7 positive activators required for the HSR and 52 negative regulators whose knockdown leads to constitutive activation of the HSR. These modifiers function in specific steps of gene expression, protein synthesis, protein folding, trafficking, and protein clearance, and comprise the metazoan heat shock regulatory network (HSN). Whereas the positive regulators function in all tissues of C. elegans, nearly all of the negative regulators exhibited tissue-selective effects. Knockdown of the subunits of the proteasome strongly induces HS reporter expression only in the intestine and spermatheca but not in muscle cells, while knockdown of subunits of the TRiC/CCT chaperonin induces HS reporter expression only in muscle cells. Yet, both the proteasome and TRiC/CCT chaperonin are ubiquitously expressed and are required for clearance and folding in all tissues. We propose that the HSN identifies a key subset of the proteostasis machinery that regulates the HSR according to the unique functional requirements of each tissue.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Cellular Proteomes Drive Tissue-Specific Regulation of the Heat Shock Response.G3 (Bethesda). 2017 Mar 10;7(3):1011-1018. doi: 10.1534/g3.116.038232. G3 (Bethesda). 2017. PMID: 28143946 Free PMC article.
-
CCAR-1 is a negative regulator of the heat-shock response in Caenorhabditis elegans.Aging Cell. 2018 Oct;17(5):e12813. doi: 10.1111/acel.12813. Epub 2018 Jul 12. Aging Cell. 2018. PMID: 30003683 Free PMC article.
-
The genome-wide role of HSF-1 in the regulation of gene expression in Caenorhabditis elegans.BMC Genomics. 2016 Aug 5;17:559. doi: 10.1186/s12864-016-2837-5. BMC Genomics. 2016. PMID: 27496166 Free PMC article.
-
Transcellular chaperone signaling: an organismal strategy for integrated cell stress responses.J Exp Biol. 2014 Jan 1;217(Pt 1):129-36. doi: 10.1242/jeb.091249. J Exp Biol. 2014. PMID: 24353212 Free PMC article. Review.
-
The functions and regulation of heat shock proteins; key orchestrators of proteostasis and the heat shock response.Arch Toxicol. 2021 Jun;95(6):1943-1970. doi: 10.1007/s00204-021-03070-8. Epub 2021 May 18. Arch Toxicol. 2021. PMID: 34003342 Review.
Cited by
-
Genetic selection for constitutively trimerized human HSF1 mutants identifies a role for coiled-coil motifs in DNA binding.G3 (Bethesda). 2013 Aug 7;3(8):1315-24. doi: 10.1534/g3.113.006692. G3 (Bethesda). 2013. PMID: 23733891 Free PMC article.
-
daf-41/p23: A Small Protein Heating Up Lifespan Regulation.PLoS Genet. 2015 Jul 6;11(7):e1005188. doi: 10.1371/journal.pgen.1005188. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26147370 Free PMC article. No abstract available.
-
MDT-15/MED15 permits longevity at low temperature via enhancing lipidostasis and proteostasis.PLoS Biol. 2019 Aug 13;17(8):e3000415. doi: 10.1371/journal.pbio.3000415. eCollection 2019 Aug. PLoS Biol. 2019. PMID: 31408455 Free PMC article.
-
BuShen HuoXue decoction improves fertility through intestinal hsp-16.2-mediated heat-shock signaling pathway in Caenorhabditis elegans.Front Pharmacol. 2023 Jun 2;14:1210701. doi: 10.3389/fphar.2023.1210701. eCollection 2023. Front Pharmacol. 2023. PMID: 37332356 Free PMC article.
-
Binding of the HSF-1 DNA-binding domain to multimeric C. elegans consensus HSEs is guided by cooperative interactions.Sci Rep. 2022 May 28;12(1):8984. doi: 10.1038/s41598-022-12736-x. Sci Rep. 2022. PMID: 35643773 Free PMC article.
References
-
- Craig EA, Gross CA (1991) Is hsp70 the cellular thermometer? Trends Biochem Sci 16: 135–140. - PubMed
-
- Abravaya K, Myers MP, Murphy SP, Morimoto RI (1992) The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev 6: 1153–1164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 GM081892/GM/NIGMS NIH HHS/United States
- R37 GM038109/GM/NIGMS NIH HHS/United States
- P40 OD010440/OD/NIH HHS/United States
- R37 AG026647/AG/NIA NIH HHS/United States
- R01 GM038109/GM/NIGMS NIH HHS/United States
- P30 CA060553/CA/NCI NIH HHS/United States
- R01 AG026647/AG/NIA NIH HHS/United States
- Z01 AG000260/ImNIH/Intramural NIH HHS/United States
- R21 NS056337/NS/NINDS NIH HHS/United States
- NIH-P50-GM081892/GM/NIGMS NIH HHS/United States
- NIH-TG-AG000260-10S1/AG/NIA NIH HHS/United States
- T32 AG000260/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

