The G4 genome
- PMID: 23637633
- PMCID: PMC3630100
- DOI: 10.1371/journal.pgen.1003468
The G4 genome
Abstract
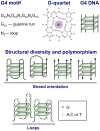
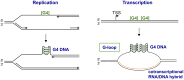
Recent experiments provide fascinating examples of how G4 DNA and G4 RNA structures--aka quadruplexes--may contribute to normal biology and to genomic pathologies. Quadruplexes are transient and therefore difficult to identify directly in living cells, which initially caused skepticism regarding not only their biological relevance but even their existence. There is now compelling evidence for functions of some G4 motifs and the corresponding quadruplexes in essential processes, including initiation of DNA replication, telomere maintenance, regulated recombination in immune evasion and the immune response, control of gene expression, and genetic and epigenetic instability. Recognition and resolution of quadruplex structures is therefore an essential component of genome biology. We propose that G4 motifs and structures that participate in key processes compose the G4 genome, analogous to the transcriptome, proteome, or metabolome. This is a new view of the genome, which sees DNA as not only a simple alphabet but also a more complex geography. The challenge for the future is to systematically identify the G4 motifs that form quadruplexes in living cells and the features that confer on specific G4 motifs the ability to function as structural elements.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

