Genetic and biochemical assays reveal a key role for replication restart proteins in group II intron retrohoming
- PMID: 23637634
- PMCID: PMC3636086
- DOI: 10.1371/journal.pgen.1003469
Genetic and biochemical assays reveal a key role for replication restart proteins in group II intron retrohoming
Abstract
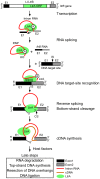
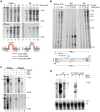
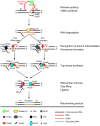
Mobile group II introns retrohome by an RNP-based mechanism in which the intron RNA reverse splices into a DNA site and is reverse transcribed by the associated intron-encoded protein. The resulting intron cDNA is then integrated into the genome by cellular mechanisms that have remained unclear. Here, we used an Escherichia coli genetic screen and Taqman qPCR assay that mitigate indirect effects to identify host factors that function in retrohoming. We then analyzed mutants identified in these and previous genetic screens by using a new biochemical assay that combines group II intron RNPs with cellular extracts to reconstitute the complete retrohoming reaction in vitro. The genetic and biochemical analyses indicate a retrohoming pathway involving degradation of the intron RNA template by a host RNase H and second-strand DNA synthesis by the host replicative DNA polymerase. Our results reveal ATP-dependent steps in both cDNA and second-strand synthesis and a surprising role for replication restart proteins in initiating second-strand synthesis in the absence of DNA replication. We also find an unsuspected requirement for host factors in initiating reverse transcription and a new RNA degradation pathway that suppresses retrohoming. Key features of the retrohoming mechanism may be used by human LINEs and other non-LTR-retrotransposons, which are related evolutionarily to mobile group II introns. Our findings highlight a new role for replication restart proteins, which function not only to repair DNA damage caused by mobile element insertion, but have also been co-opted to become an integral part of the group II intron retrohoming mechanism.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The retrohoming of linear group II intron RNAs in Drosophila melanogaster occurs by both DNA ligase 4-dependent and -independent mechanisms.PLoS Genet. 2012;8(2):e1002534. doi: 10.1371/journal.pgen.1002534. Epub 2012 Feb 16. PLoS Genet. 2012. PMID: 22359518 Free PMC article.
-
Recruitment of host functions suggests a repair pathway for late steps in group II intron retrohoming.Genes Dev. 2005 Oct 15;19(20):2477-87. doi: 10.1101/gad.1345105. Genes Dev. 2005. PMID: 16230535 Free PMC article.
-
Retrohoming of a Mobile Group II Intron in Human Cells Suggests How Eukaryotes Limit Group II Intron Proliferation.PLoS Genet. 2015 Aug 4;11(8):e1005422. doi: 10.1371/journal.pgen.1005422. eCollection 2015 Aug. PLoS Genet. 2015. PMID: 26241656 Free PMC article.
-
Mobile group II introns.Annu Rev Genet. 2004;38:1-35. doi: 10.1146/annurev.genet.38.072902.091600. Annu Rev Genet. 2004. PMID: 15568970 Review.
-
Group II introns and expression of conjugative transfer functions in lactic acid bacteria.Antonie Van Leeuwenhoek. 1999 Jul-Nov;76(1-4):77-88. Antonie Van Leeuwenhoek. 1999. PMID: 10532373 Review.
Cited by
-
Group II intron-ribosome association protects intron RNA from degradation.RNA. 2013 Nov;19(11):1497-509. doi: 10.1261/rna.039073.113. Epub 2013 Sep 17. RNA. 2013. PMID: 24046482 Free PMC article.
-
Integration, Regulation, and Long-Term Stability of R2 Retrotransposons.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0011-2014. doi: 10.1128/microbiolspec.MDNA3-0011-2014. Microbiol Spectr. 2015. PMID: 26104703 Free PMC article. Review.
-
A targetron system for gene targeting in thermophiles and its application in Clostridium thermocellum.PLoS One. 2013 Jul 9;8(7):e69032. doi: 10.1371/journal.pone.0069032. Print 2013. PLoS One. 2013. PMID: 23874856 Free PMC article.
-
Functionality of In vitro Reconstituted Group II Intron RmInt1-Derived Ribonucleoprotein Particles.Front Mol Biosci. 2016 Sep 27;3:58. doi: 10.3389/fmolb.2016.00058. eCollection 2016. Front Mol Biosci. 2016. PMID: 27730127 Free PMC article.
-
Mobile Bacterial Group II Introns at the Crux of Eukaryotic Evolution.Microbiol Spectr. 2015 Feb;3(1):MDNA3-0050-2014. doi: 10.1128/microbiolspec.MDNA3-0050-2014. Microbiol Spectr. 2015. PMID: 26104554 Free PMC article. Review.
References
-
- Lambowitz AM, Zimmerly S (2011) Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol 3: a003616 doi:10.1101/cshperspect.a003616. - DOI - PMC - PubMed
-
- Zimmerly S, Guo H, Eskes R, Yang J, Perlman PS, et al. (1995) A group II intron RNA is a catalytic component of a DNA endonuclease involved in intron mobility. Cell 83: 529–538. - PubMed
-
- Zimmerly S, Guo H, Perlman PS, Lambowitz AM (1995) Group II intron mobility occurs by target DNA-primed reverse transcription. Cell 82: 545–554. - PubMed
-
- Yang J, Zimmerly S, Perlman PS, Lambowitz AM (1996) Efficient integration of an intron RNA into double-stranded DNA by reverse splicing. Nature 381: 332–335. - PubMed
-
- Cousineau B, Smith D, Lawrence-Cavanagh S, Mueller JE, Yang J, et al. (1998) Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell 94: 451–462. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

