From data towards knowledge: revealing the architecture of signaling systems by unifying knowledge mining and data mining of systematic perturbation data
- PMID: 23637789
- PMCID: PMC3634064
- DOI: 10.1371/journal.pone.0061134
From data towards knowledge: revealing the architecture of signaling systems by unifying knowledge mining and data mining of systematic perturbation data
Abstract
Genetic and pharmacological perturbation experiments, such as deleting a gene and monitoring gene expression responses, are powerful tools for studying cellular signal transduction pathways. However, it remains a challenge to automatically derive knowledge of a cellular signaling system at a conceptual level from systematic perturbation-response data. In this study, we explored a framework that unifies knowledge mining and data mining towards the goal. The framework consists of the following automated processes: 1) applying an ontology-driven knowledge mining approach to identify functional modules among the genes responding to a perturbation in order to reveal potential signals affected by the perturbation; 2) applying a graph-based data mining approach to search for perturbations that affect a common signal; and 3) revealing the architecture of a signaling system by organizing signaling units into a hierarchy based on their relationships. Applying this framework to a compendium of yeast perturbation-response data, we have successfully recovered many well-known signal transduction pathways; in addition, our analysis has led to many new hypotheses regarding the yeast signal transduction system; finally, our analysis automatically organized perturbed genes as a graph reflecting the architecture of the yeast signaling system. Importantly, this framework transformed molecular findings from a gene level to a conceptual level, which can be readily translated into computable knowledge in the form of rules regarding the yeast signaling system, such as "if genes involved in the MAPK signaling are perturbed, genes involved in pheromone responses will be differentially expressed."
Conflict of interest statement
Figures

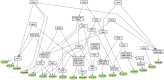
 represents the root of the Biological Process namespace.
represents the root of the Biological Process namespace.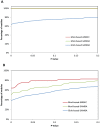





References
-
- Cover TM, Thomas JA (2006) Elements of Information Theory. 2nd Ed., John Wiley and Sons.
-
- Herskowitz I (1995) MAP kinase pathways in yeast: For mating and more. Cell 80: 187–197. - PubMed
-
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, et al. (2000) Functional Discovery via a Compendium of Expression Profiles. Cell 102: 109–126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

