Clostridium botulinum strain Af84 contains three neurotoxin gene clusters: bont/A2, bont/F4 and bont/F5
- PMID: 23637798
- PMCID: PMC3625220
- DOI: 10.1371/journal.pone.0061205
Clostridium botulinum strain Af84 contains three neurotoxin gene clusters: bont/A2, bont/F4 and bont/F5
Abstract
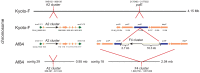
Sanger and shotgun sequencing of Clostridium botulinum strain Af84 type Af and its botulinum neurotoxin gene (bont) clusters identified the presence of three bont gene clusters rather than the expected two. The three toxin gene clusters consisted of bont subtypes A2, F4 and F5. The bont/A2 and bont/F4 gene clusters were located within the chromosome (the latter in a novel location), while the bont/F5 toxin gene cluster was located within a large 246 kb plasmid. These findings are the first identification of a C. botulinum strain that contains three botulinum neurotoxin gene clusters.
Conflict of interest statement
Figures



References
-
- Arnon SS, Schechter R, Inglesby T, Henderson D, Bertlett J, et al. (2001) Botulinum toxin as a biological weapon: medical and public health management. JAMA 285: 1059–1070. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

