Identification of genes and pathways related to phenol degradation in metagenomic libraries from petroleum refinery wastewater
- PMID: 23637911
- PMCID: PMC3630121
- DOI: 10.1371/journal.pone.0061811
Identification of genes and pathways related to phenol degradation in metagenomic libraries from petroleum refinery wastewater
Abstract
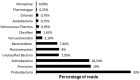
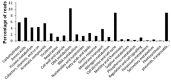
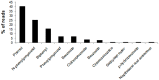
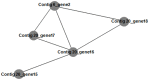
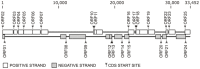
Two fosmid libraries, totaling 13,200 clones, were obtained from bioreactor sludge of petroleum refinery wastewater treatment system. The library screening based on PCR and biological activity assays revealed more than 400 positive clones for phenol degradation. From these, 100 clones were randomly selected for pyrosequencing in order to evaluate the genetic potential of the microorganisms present in wastewater treatment plant for biodegradation, focusing mainly on novel genes and pathways of phenol and aromatic compound degradation. The sequence analysis of selected clones yielded 129,635 reads at an estimated 17-fold coverage. The phylogenetic analysis showed Burkholderiales and Rhodocyclales as the most abundant orders among the selected fosmid clones. The MG-RAST analysis revealed a broad metabolic profile with important functions for wastewater treatment, including metabolism of aromatic compounds, nitrogen, sulphur and phosphorus. The predicted 2,276 proteins included phenol hydroxylases and cathecol 2,3- dioxygenases, involved in the catabolism of aromatic compounds, such as phenol, byphenol, benzoate and phenylpropanoid. The sequencing of one fosmid insert of 33 kb unraveled the gene that permitted the host, Escherichia coli EPI300, to grow in the presence of aromatic compounds. Additionally, the comparison of the whole fosmid sequence against bacterial genomes deposited in GenBank showed that about 90% of sequence showed no identity to known sequences of Proteobacteria deposited in the NCBI database. This study surveyed the functional potential of fosmid clones for aromatic compound degradation and contributed to our knowledge of the biodegradative capacity and pathways of microbial assemblages present in refinery wastewater treatment system.
Conflict of interest statement
Figures

References
-
- Barrios-Martinez A, Barbot E, Marrot B, Moulin P, Roche N (2006) Degradation of synthetic phenol-containing wastewaters by MBR. J Membrane Sci 281: 288–296.
-
- Cordova-Rosa SM, Dams RI, Cordova-Rosa EV, Radetski MR, Corrêa AXR, et al. (2009) Remediation of phenol-contaminated soil by a bacterial consortium and Acinetobacter calcoaceticus isolated from an industrial wastewater treatment plant. J Hazard Mater 164: 61–66. - PubMed
-
- Dong X, Hong Q, He L, Jiang X, Li S (2008) Characterization of phenol-degrading bacterial strains isolated from natural soil. Inter Biodet Biodeg 62: 257–262.
-
- Valle A, Bailey MJ, Whiteley AS, Manefield M (2004) N -acyl- L -homoserine lactones (AHLs) affect microbial community composition and function in activated sludge. Environ Microbiol 6: 424–433. - PubMed
-
- Nair IC, Jayachandran K, Shashidhar S (2007) Treatment of paper factory effluent using a phenol degrading Alcaligenes sp. under free and immobilized conditions. Biores Technol 98: 714–716. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

