Bioinformatics analysis identify novel OB fold protein coding genes in C. elegans
- PMID: 23638006
- PMCID: PMC3636199
- DOI: 10.1371/journal.pone.0062204
Bioinformatics analysis identify novel OB fold protein coding genes in C. elegans
Abstract
Background: The C. elegans genome has been extensively annotated by the WormBase consortium that uses state of the art bioinformatics pipelines, functional genomics and manual curation approaches. As a result, the identification of novel genes in silico in this model organism is becoming more challenging requiring new approaches. The Oligonucleotide-oligosaccharide binding (OB) fold is a highly divergent protein family, in which protein sequences, in spite of having the same fold, share very little sequence identity (5-25%). Therefore, evidence from sequence-based annotation may not be sufficient to identify all the members of this family. In C. elegans, the number of OB-fold proteins reported is remarkably low (n=46) compared to other evolutionary-related eukaryotes, such as yeast S. cerevisiae (n=344) or fruit fly D. melanogaster (n=84). Gene loss during evolution or differences in the level of annotation for this protein family, may explain these discrepancies.
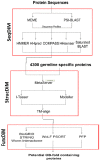
Methodology/principal findings: This study examines the possibility that novel OB-fold coding genes exist in the worm. We developed a bioinformatics approach that uses the most sensitive sequence-sequence, sequence-profile and profile-profile similarity search methods followed by 3D-structure prediction as a filtering step to eliminate false positive candidate sequences. We have predicted 18 coding genes containing the OB-fold that have remarkably partially been characterized in C. elegans.
Conclusions/significance: This study raises the possibility that the annotation of highly divergent protein fold families can be improved in C. elegans. Similar strategies could be implemented for large scale analysis by the WormBase consortium when novel versions of the genome sequence of C. elegans, or other evolutionary related species are being released. This approach is of general interest to the scientific community since it can be used to annotate any genome.
Conflict of interest statement
Figures

Similar articles
-
WormBase: a comprehensive data resource for Caenorhabditis biology and genomics.Nucleic Acids Res. 2005 Jan 1;33(Database issue):D383-9. doi: 10.1093/nar/gki066. Nucleic Acids Res. 2005. PMID: 15608221 Free PMC article.
-
NemaFootPrinter: a web based software for the identification of conserved non-coding genome sequence regions between C. elegans and C. briggsae.BMC Bioinformatics. 2005 Dec 1;6 Suppl 4(Suppl 4):S22. doi: 10.1186/1471-2105-6-S4-S22. BMC Bioinformatics. 2005. PMID: 16351749 Free PMC article.
-
RUN-CBFbeta interaction in C. elegans: computational prediction and experimental verification.J Biomol Struct Dyn. 2007 Feb;24(4):343-58. doi: 10.1080/07391102.2007.10507124. J Biomol Struct Dyn. 2007. PMID: 17206850
-
Functional genomic approaches using the nematode Caenorhabditis elegans as a model system.J Biochem Mol Biol. 2004 Jan 31;37(1):107-13. doi: 10.5483/bmbrep.2004.37.1.107. J Biochem Mol Biol. 2004. PMID: 14761308 Review.
-
The Remarkably Diverse Family of T-Box Factors in Caenorhabditis elegans.Curr Top Dev Biol. 2017;122:27-54. doi: 10.1016/bs.ctdb.2016.08.005. Epub 2016 Sep 21. Curr Top Dev Biol. 2017. PMID: 28057267 Review.
Cited by
-
Exploring the structural landscape of DNA maintenance proteins.Nat Commun. 2024 Sep 5;15(1):7748. doi: 10.1038/s41467-024-49983-7. Nat Commun. 2024. PMID: 39237506 Free PMC article.
-
AnABlast: a new in silico strategy for the genome-wide search of novel genes and fossil regions.DNA Res. 2015 Dec;22(6):439-49. doi: 10.1093/dnares/dsv025. Epub 2015 Oct 21. DNA Res. 2015. PMID: 26494834 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

